
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,231,589 – 20,231,717 |
| Length | 128 |
| Max. P | 0.664798 |

| Location | 20,231,589 – 20,231,717 |
|---|---|
| Length | 128 |
| Sequences | 5 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 78.96 |
| Shannon entropy | 0.35959 |
| G+C content | 0.42624 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -17.68 |
| Energy contribution | -19.12 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
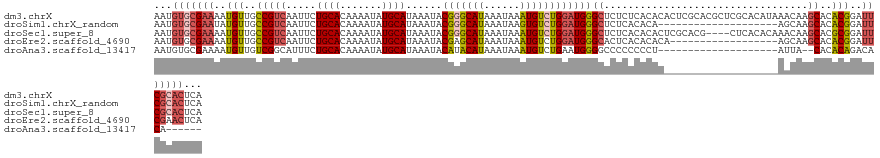
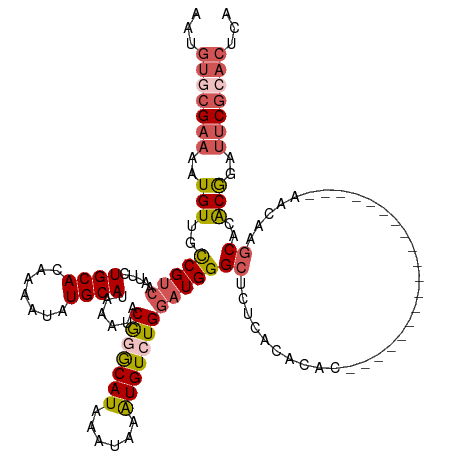
>dm3.chrX 20231589 128 + 22422827 AAUGUGCGAAAAUGUUGCCGUCAAUUCUGCACAAAAUAUGCAUAAAUACGGGCAUAAAUAAAUGUCUGGAUGGGCUCUCUCACACACUCGCACGCUCGCACAUAAACAAGCACACGGAUUCGCACUCA ...(((((((..((((((((((.....((((.......))))......(((((((......))))))))))).................((......))..........))).)))..)))))))... ( -30.70, z-score = -1.47, R) >droSim1.chrX_random 5260781 108 + 5698898 AAUGUGCGAAUAUGUUGCCGUCAAUUCUGCACAAAAUAUGCAUAAAUACGGGCAUAAAUAAGUGUCUGGAUGGGCUCUCACACA--------------------AGCAAGCACACGGAUUCGCACUCA ...((((((((.((((((((((.....((((.......))))......(((((((......))))))))))).(((........--------------------)))..))).))).))))))))... ( -32.20, z-score = -2.51, R) >droSec1.super_8 2419827 124 + 3762037 AAUGUGCGAAAAUGUUGCCGUCAAUUCUGCACAAAAUAUGCAUAAAUACGGGCAUAAAUAAAUGUCUGGAUGGGCUCUCACACACUCGCACG----CUCACACAAACAAGCACGCGGAUUCGCACUCA ...(((((((.......(((((.....((((.......))))......(((((((......))))))))))))............((((..(----((..........)))..)))).)))))))... ( -32.20, z-score = -1.92, R) >droEre2.scaffold_4690 8430442 110 - 18748788 AAUGUGCGAAAAUGUUGCCGUCAAUUCUGCACAAAAUAUGCAUAAAUACGAGCAUAAAUAAAUGUCUGGAUGGGCACUCACACACA------------------AGCAAGCACACGGAUUCGAACUCA ..(((((........((((((...((.((((.......)))).))..))).)))........((..((..((........))..))------------------..)).))))).............. ( -19.60, z-score = 0.82, R) >droAna3.scaffold_13417 5436330 100 - 6960332 AAUGUGCGAAAAUGUUGUCGGCAUUUCUGCACAAAAUAUGCAUAAAUACAUACAUAAAUAAAUGUCUGAAUGGGGCCCCCCCCU--------------------AUUA--CACACAGACACA------ ..((((((.((((((.....)))))).))))))...((((........))))..........(((((((((((((.....))))--------------------))).--....))))))..------ ( -26.40, z-score = -2.64, R) >consensus AAUGUGCGAAAAUGUUGCCGUCAAUUCUGCACAAAAUAUGCAUAAAUACGGGCAUAAAUAAAUGUCUGGAUGGGCUCUCACACAC___________________AACAAGCACACGGAUUCGCACUCA ...(((((((..(((..(((((.....((((.......))))......(((((((......))))))))))))((..................................))..)))..)))))))... (-17.68 = -19.12 + 1.44)
| Location | 20,231,589 – 20,231,717 |
|---|---|
| Length | 128 |
| Sequences | 5 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 78.96 |
| Shannon entropy | 0.35959 |
| G+C content | 0.42624 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -18.16 |
| Energy contribution | -19.24 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
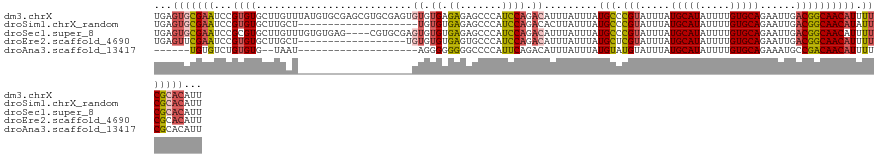
>dm3.chrX 20231589 128 - 22422827 UGAGUGCGAAUCCGUGUGCUUGUUUAUGUGCGAGCGUGCGAGUGUGUGAGAGAGCCCAUCCAGACAUUUAUUUAUGCCCGUAUUUAUGCAUAUUUUGUGCAGAAUUGACGGCAACAUUUUCGCACAUU ...(((((((...(..(((((((......)))))))..)((((((.((.((.......)))).)))))).....((((.((.....(((((.....)))))......)))))).....)))))))... ( -43.10, z-score = -3.22, R) >droSim1.chrX_random 5260781 108 - 5698898 UGAGUGCGAAUCCGUGUGCUUGCU--------------------UGUGUGAGAGCCCAUCCAGACACUUAUUUAUGCCCGUAUUUAUGCAUAUUUUGUGCAGAAUUGACGGCAACAUAUUCGCACAUU ...((((((((..(((((..((..--------------------((.((....)).))..))..))).......((((.((.....(((((.....)))))......))))))))..))))))))... ( -32.40, z-score = -2.48, R) >droSec1.super_8 2419827 124 - 3762037 UGAGUGCGAAUCCGCGUGCUUGUUUGUGUGAG----CGUGCGAGUGUGUGAGAGCCCAUCCAGACAUUUAUUUAUGCCCGUAUUUAUGCAUAUUUUGUGCAGAAUUGACGGCAACAUUUUCGCACAUU ...(((((((...(((((((..(....)..))----)))))((((((.((.((.....)))).)))))).....((((.((.....(((((.....)))))......)))))).....)))))))... ( -40.50, z-score = -2.60, R) >droEre2.scaffold_4690 8430442 110 + 18748788 UGAGUUCGAAUCCGUGUGCUUGCU------------------UGUGUGUGAGUGCCCAUCCAGACAUUUAUUUAUGCUCGUAUUUAUGCAUAUUUUGUGCAGAAUUGACGGCAACAUUUUCGCACAUU ...((.((((...(.(..((..(.------------------.....)..))..).).................(((.(((.....(((((.....)))))......)))))).....)))).))... ( -25.20, z-score = 0.08, R) >droAna3.scaffold_13417 5436330 100 + 6960332 ------UGUGUCUGUGUG--UAAU--------------------AGGGGGGGGCCCCAUUCAGACAUUUAUUUAUGUAUGUAUUUAUGCAUAUUUUGUGCAGAAAUGCCGACAACAUUUUCGCACAUU ------.(((((((....--.(((--------------------.((((....)))))))))))))).....((((((((....))))))))...(((((..(((((.......)))))..))))).. ( -30.20, z-score = -1.92, R) >consensus UGAGUGCGAAUCCGUGUGCUUGCU___________________GUGUGUGAGAGCCCAUCCAGACAUUUAUUUAUGCCCGUAUUUAUGCAUAUUUUGUGCAGAAUUGACGGCAACAUUUUCGCACAUU ...(((((((...((((..........................((.((.((.......)))).)).........(((.(((.....(((((.....)))))......)))))))))).)))))))... (-18.16 = -19.24 + 1.08)
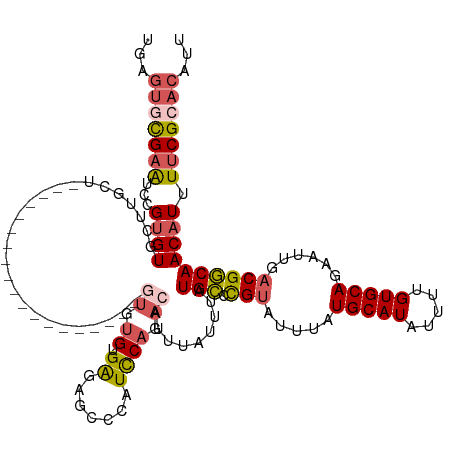
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:26 2011