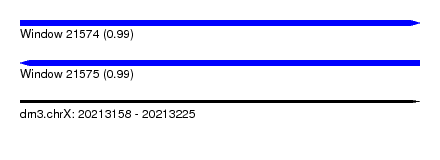
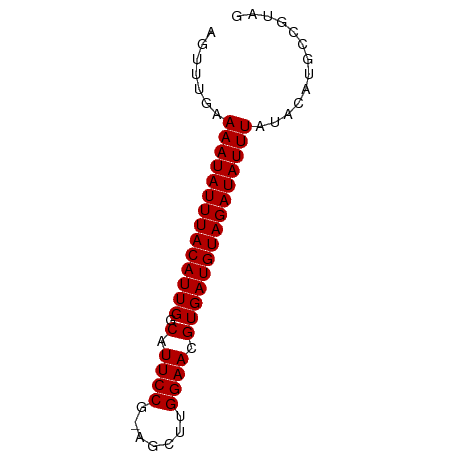
| Sequence ID | dm3.chrX |
|---|---|
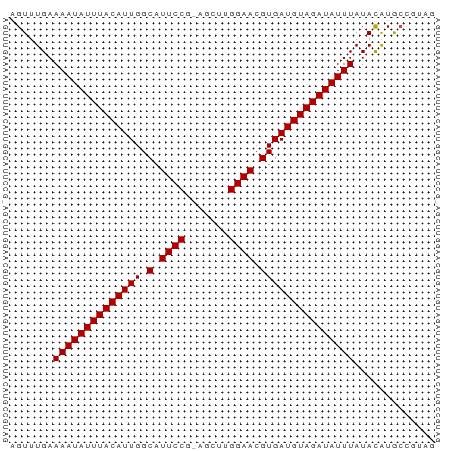
| Location | 20,213,158 – 20,213,225 |
| Length | 67 |
| Max. P | 0.994146 |

| Location | 20,213,158 – 20,213,225 |
|---|---|
| Length | 67 |
| Sequences | 11 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 90.72 |
| Shannon entropy | 0.20405 |
| G+C content | 0.36208 |
| Mean single sequence MFE | -13.26 |
| Consensus MFE | -12.47 |
| Energy contribution | -12.65 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20213158 67 + 22422827 CUACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCU-CGGAAUGCCAAUGUAAAUAUUUUCAAACU ........((...((((((.(((((..((((((.....-.))))))...))))).)))))).)).... ( -13.50, z-score = -3.04, R) >droSec1.super_8 2398112 67 + 3762037 CUACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCU-CGGAAUGCCAAUGUAAAUAUUUUCAAACU ........((...((((((.(((((..((((((.....-.))))))...))))).)))))).)).... ( -13.50, z-score = -3.04, R) >droYak2.chrX 19442994 67 + 21770863 CUACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCU-CGGAAUGCCAAUGUAAAUAUUUUCAAACU ........((...((((((.(((((..((((((.....-.))))))...))))).)))))).)).... ( -13.50, z-score = -3.04, R) >droEre2.scaffold_4690 10290722 67 + 18748788 CUACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCU-CGGAAUGCCAAUGUAAAUAUUUUCAAACU ........((...((((((.(((((..((((((.....-.))))))...))))).)))))).)).... ( -13.50, z-score = -3.04, R) >droAna3.scaffold_13417 5412963 66 - 6960332 --ACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCUUCGGAAUGCCAAUGUAAAUAUUUUCAAACU --......((...((((((.(((((..((((((.......))))))...))))).)))))).)).... ( -13.00, z-score = -2.69, R) >dp4.chrXL_group1e 5466647 67 - 12523060 CUACAGCAUGUAUAAAUAUCUACAUCACGUUCCAGGGC-CGGAAUGCCAAUGUAAAUAUUUUCAAACU ........((...((((((.(((((..((((((.....-.))))))...))))).)))))).)).... ( -13.50, z-score = -2.16, R) >droPer1.super_13 1497432 67 + 2293547 CUACAGCAUGUAUAAAUAUCUACAUCACGUUCCAGGGC-CGGAAUGCCAAUGUAAAUAUUUUCAAACU ........((...((((((.(((((..((((((.....-.))))))...))))).)))))).)).... ( -13.50, z-score = -2.16, R) >droWil1.scaffold_181096 9264873 67 + 12416693 CUCGGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCU-CGGAAUGCCAAUGUAAAUAUUUUCAAACU ........((...((((((.(((((..((((((.....-.))))))...))))).)))))).)).... ( -13.50, z-score = -2.62, R) >droMoj3.scaffold_6308 523541 66 + 3356042 --CAUGUACAUAUAAAUAUCUACAUCACGUUCCGAGCUCGGGAAUGCCAAUGUAAAUAUUUUCAAACU --...........((((((.(((((..((((((.......))))))...))))).))))))....... ( -12.20, z-score = -2.19, R) >droVir3.scaffold_13042 4569291 66 - 5191987 --CUGCCAUGUAUAAAUAUCUACAUCACGUUCCGAGCUCGGGAAUGCCAAUGUAAAUAUUUUCAAACU --......((...((((((.(((((..((((((.......))))))...))))).)))))).)).... ( -13.10, z-score = -2.10, R) >droGri2.scaffold_15203 6066532 62 - 11997470 ------CAUGUAUAAAUAUCUACAUCACGUUCCGUGCUCGGGAAUGCCAAUGUAAAUAUUUUCAAACU ------..((...((((((.(((((..((((((.......))))))...))))).)))))).)).... ( -13.10, z-score = -2.70, R) >consensus CUACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCU_CGGAAUGCCAAUGUAAAUAUUUUCAAACU ........((...((((((.(((((..((((((.......))))))...))))).)))))).)).... (-12.47 = -12.65 + 0.18)

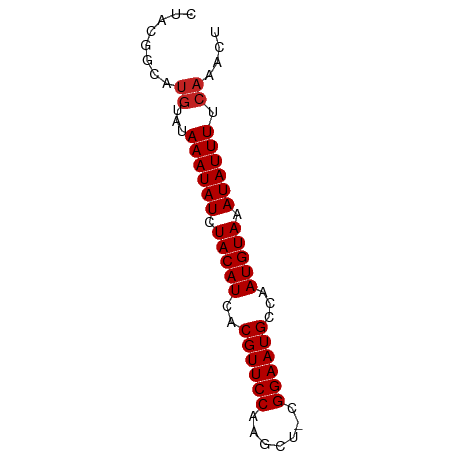
| Location | 20,213,158 – 20,213,225 |
|---|---|
| Length | 67 |
| Sequences | 11 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 90.72 |
| Shannon entropy | 0.20405 |
| G+C content | 0.36208 |
| Mean single sequence MFE | -15.85 |
| Consensus MFE | -14.75 |
| Energy contribution | -14.75 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20213158 67 - 22422827 AGUUUGAAAAUAUUUACAUUGGCAUUCCG-AGCUUGGAACGUGAUGUAGAUAUUUAUACAUGCCGUAG .......((((((((((((((.(.(((((-....))))).)))))))))))))))............. ( -15.70, z-score = -2.20, R) >droSec1.super_8 2398112 67 - 3762037 AGUUUGAAAAUAUUUACAUUGGCAUUCCG-AGCUUGGAACGUGAUGUAGAUAUUUAUACAUGCCGUAG .......((((((((((((((.(.(((((-....))))).)))))))))))))))............. ( -15.70, z-score = -2.20, R) >droYak2.chrX 19442994 67 - 21770863 AGUUUGAAAAUAUUUACAUUGGCAUUCCG-AGCUUGGAACGUGAUGUAGAUAUUUAUACAUGCCGUAG .......((((((((((((((.(.(((((-....))))).)))))))))))))))............. ( -15.70, z-score = -2.20, R) >droEre2.scaffold_4690 10290722 67 - 18748788 AGUUUGAAAAUAUUUACAUUGGCAUUCCG-AGCUUGGAACGUGAUGUAGAUAUUUAUACAUGCCGUAG .......((((((((((((((.(.(((((-....))))).)))))))))))))))............. ( -15.70, z-score = -2.20, R) >droAna3.scaffold_13417 5412963 66 + 6960332 AGUUUGAAAAUAUUUACAUUGGCAUUCCGAAGCUUGGAACGUGAUGUAGAUAUUUAUACAUGCCGU-- .......((((((((((((((.(.((((((...)))))).)))))))))))))))...........-- ( -16.10, z-score = -2.39, R) >dp4.chrXL_group1e 5466647 67 + 12523060 AGUUUGAAAAUAUUUACAUUGGCAUUCCG-GCCCUGGAACGUGAUGUAGAUAUUUAUACAUGCUGUAG (((.((.((((((((((((((.(.(((((-....))))).)))))))))))))))...)).))).... ( -17.80, z-score = -2.49, R) >droPer1.super_13 1497432 67 - 2293547 AGUUUGAAAAUAUUUACAUUGGCAUUCCG-GCCCUGGAACGUGAUGUAGAUAUUUAUACAUGCUGUAG (((.((.((((((((((((((.(.(((((-....))))).)))))))))))))))...)).))).... ( -17.80, z-score = -2.49, R) >droWil1.scaffold_181096 9264873 67 - 12416693 AGUUUGAAAAUAUUUACAUUGGCAUUCCG-AGCUUGGAACGUGAUGUAGAUAUUUAUACAUGCCCGAG .......((((((((((((((.(.(((((-....))))).)))))))))))))))............. ( -15.70, z-score = -2.36, R) >droMoj3.scaffold_6308 523541 66 - 3356042 AGUUUGAAAAUAUUUACAUUGGCAUUCCCGAGCUCGGAACGUGAUGUAGAUAUUUAUAUGUACAUG-- .((.((.((((((((((((((.(.((((.(....))))).))))))))))))))).))...))...-- ( -14.90, z-score = -1.91, R) >droVir3.scaffold_13042 4569291 66 + 5191987 AGUUUGAAAAUAUUUACAUUGGCAUUCCCGAGCUCGGAACGUGAUGUAGAUAUUUAUACAUGGCAG-- .......((((((((((((((.(.((((.(....))))).)))))))))))))))...........-- ( -14.70, z-score = -1.61, R) >droGri2.scaffold_15203 6066532 62 + 11997470 AGUUUGAAAAUAUUUACAUUGGCAUUCCCGAGCACGGAACGUGAUGUAGAUAUUUAUACAUG------ .......((((((((((((((.(.((((.......)))).))))))))))))))).......------ ( -14.60, z-score = -2.60, R) >consensus AGUUUGAAAAUAUUUACAUUGGCAUUCCG_AGCUUGGAACGUGAUGUAGAUAUUUAUACAUGCCGUAG .......((((((((((((((.(.((((.......)))).)))))))))))))))............. (-14.75 = -14.75 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:24 2011