| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,211,083 – 20,211,177 |
| Length | 94 |
| Max. P | 0.815205 |

| Location | 20,211,083 – 20,211,177 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.07 |
| Shannon entropy | 0.40002 |
| G+C content | 0.37203 |
| Mean single sequence MFE | -20.17 |
| Consensus MFE | -9.38 |
| Energy contribution | -9.93 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.815205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20211083 94 + 22422827 UUUAUGACACUUUGUCGUAUUGUUUUUUGUUU-----------GGCUUCUAC--CGUACUUUGUGG-CGAACGUGUCAUAUUCAGUUACGUAUUUAUUCCUUGCCCAG---------- ..(((((((((((((((((..((........(-----------((......)--)).))..)))))-)))).))))))))............................---------- ( -17.80, z-score = -1.28, R) >droSim1.chrX 15506361 94 + 17042790 UUUAUGACACUUUGUCGUAUUGUUUUUUGUUU-----------GGCUUCUAC--CGUACUUUGCGG-CGAACGUGUCAUAUUCAGUUAUGUAUUUAUUCCUUGCCCAG---------- ..(((((((((((((((((..((........(-----------((......)--)).))..)))))-)))).))))))))............................---------- ( -22.60, z-score = -3.10, R) >droSec1.super_8 2396217 94 + 3762037 UUUAUGACACUUUGUCGUAUUGUUUUUUGUUU-----------GGCUUCUAC--CGUACUUUGCGG-CGAACGUGUCAUAUUCAGUUAUGUAUUUAUUCCUUGCCCAG---------- ..(((((((((((((((((..((........(-----------((......)--)).))..)))))-)))).))))))))............................---------- ( -22.60, z-score = -3.10, R) >droAna3.scaffold_13417 5409563 107 - 6960332 UUUAUGACACUUUGUCGUAUUGUUUUCUGUUCUUUGGAGCCUCGGCUUGUGCGCCGCACUUUGCGG-CGAACGUGUCAUAUUCAGUUAUGUAUUUAUUCCUUGUCCAG---------- ..(((((((((((((((((..((.....((((....))))..((((......)))).))..)))))-)))).))))))))............................---------- ( -31.00, z-score = -3.26, R) >droWil1.scaffold_181096 3157504 82 - 12416693 UUUAUGACACUUUGUCGUAUUGUUUUGU--------------------------UGUACUUUGCGGACGAACGUGUCAUAUUCAGUUAUGUAUUUAUUUCUUGGCCAG---------- ..(((((((((((((((((..((.....--------------------------...))..))).)))))).))))))))............................---------- ( -19.60, z-score = -2.32, R) >droGri2.scaffold_15203 6064460 91 - 11997470 UUCAUGACACUUUGUCGUAUUGUUGUUU--------------------------UCUACUUUGCAAC-GAACGUGUCAUAUUCAGUUAUGUAUUUAUUCCAAUUCCACAAGCCUCUUC ...((((((((((((.(((..((.....--------------------------...))..))).))-))).)))))))....................................... ( -16.40, z-score = -2.47, R) >droMoj3.scaffold_6308 520855 86 + 3356042 UUUAUGACACUUUGUCUUAUUGUUGUUU--------------------------UCUACUUUGCAGCAGAACGUGUCAUAUUCAGUUAUGUAUUUAUUCCAAUUGCAGCCAA------ ..((((((((....(((....(((((..--------------------------........))))))))..))))))))........((((...........)))).....------ ( -14.20, z-score = -0.74, R) >droVir3.scaffold_13042 4566974 84 - 5191987 UUUAUGACACUUUGUCUUAUUGUUGUUU--------------------------UCUACUUUGCGGC-GAACGUGUCAUAUUCAGUUAUGUAUUUAUUCCAAUUGCACCGG------- ..((((((((((((((.((..((.....--------------------------...))..)).)))-))).))))))))........((((...........))))....------- ( -17.20, z-score = -2.00, R) >consensus UUUAUGACACUUUGUCGUAUUGUUUUUU__________________________CGUACUUUGCGG_CGAACGUGUCAUAUUCAGUUAUGUAUUUAUUCCUUGUCCAG__________ ..(((((((((((..((((..((..................................))..))))...))).))))))))...................................... ( -9.38 = -9.93 + 0.55)

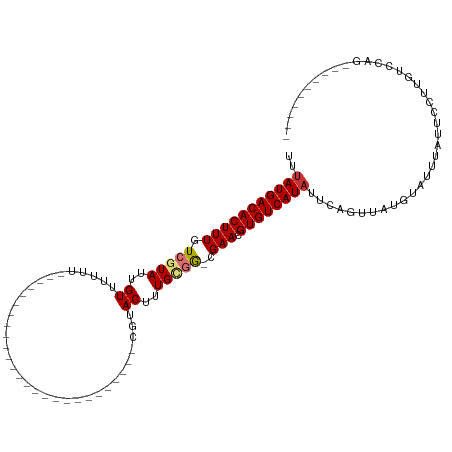
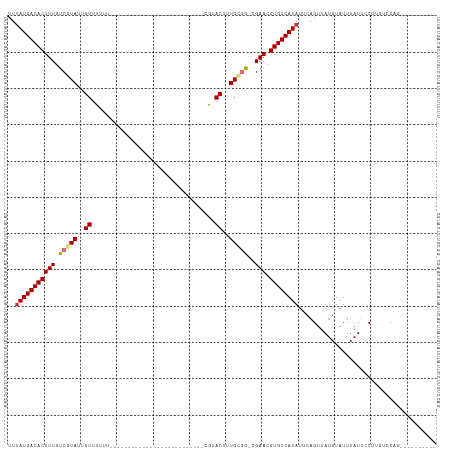
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:22 2011