| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,196,183 – 20,196,279 |
| Length | 96 |
| Max. P | 0.834629 |

| Location | 20,196,183 – 20,196,279 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.58 |
| Shannon entropy | 0.52470 |
| G+C content | 0.42327 |
| Mean single sequence MFE | -17.24 |
| Consensus MFE | -10.89 |
| Energy contribution | -11.18 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
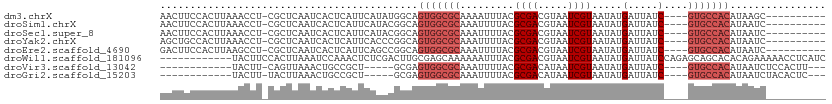
>dm3.chrX 20196183 96 - 22422827 AACUUCCACUUAAACCU-CGCUCAAUCACUCAUUCAUAUGGCAGUGGCGCAAAAUUUACGCGACGUAAUCGUAAUAUGAUUAUC----GUGCCACAUAAGC---------- ........((((.....-.((.((..............)))).(((((((.........)).((((((((((...))))))).)----))))))).)))).---------- ( -18.04, z-score = -1.10, R) >droSim1.chrX 15477133 96 - 17042790 AACUUCCACUUAAACCU-CGCUCAAUCACUCAUUCAUACGGCAGUGGCGCAAAUUUUACGCGACGUAAUCGUAAUAUGAUUAUC----GUGCCACAUAAUC---------- .................-.(((.................))).(((((((.........)).((((((((((...))))))).)----)))))))......---------- ( -16.93, z-score = -1.31, R) >droSec1.super_8 2381598 96 - 3762037 AACUUCCACUUAAACCU-CGCUCAAUCACUCAUUCAUACGGCAGUGGCGCAAAUUUUACGCGACGUAAUCGUAAUAUGAUUAUC----GUGCCACAUAAUC---------- .................-.(((.................))).(((((((.........)).((((((((((...))))))).)----)))))))......---------- ( -16.93, z-score = -1.31, R) >droYak2.chrX 19425680 96 - 21770863 AGCUGCCACUUAAACCU-CGCUCAAUCACUCAUUCACCCGGCAGUGGCGCAAAUUUUACGCGACGUAAUCGUAAUAUGAUUAUC----GUGCCACAUAAUC---------- .((((((..........-.....................))))))(((((.........)).((((((((((...))))))).)----)))))........---------- ( -19.58, z-score = -1.13, R) >droEre2.scaffold_4690 10275258 96 - 18748788 GACUUCCACUUAAGCCU-CGCUCAAUCACUCAUUCAGCCGGCAGUGGCGCAAAUUUUACGCGACGUAAUCGUAAUAUGAUUAUC----GUGCCACAUAAUC---------- .............(((.-.(((.(((.....))).))).))).(((((((.........)).((((((((((...))))))).)----)))))))......---------- ( -23.40, z-score = -2.37, R) >droWil1.scaffold_181096 9243749 99 - 12416693 ------------UACUUCCACUUAAAUCCAAACUCUCGACUUGCGAGCAAAAAAUUUACGCGACGUAAUCGUAAUAUGAUUAUCCAGAGCAGCACACAGAAAAACCUCAUC ------------....................((((.((.(((((.............)))))..(((((((...))))))))).))))...................... ( -10.02, z-score = 0.20, R) >droVir3.scaffold_13042 4542114 86 + 5191987 ------------UACUU-CAGUUAAACUGCCGCU-----GCGAGUGGCGCAAAUUUUACGCGACAUAAUCGUAAUAUGAUUAUC----GUGCCACAUAAUCUCCACUU--- ------------.....-..........(((((.-----....)))))..........(((((..(((((((...)))))))))----))).................--- ( -16.50, z-score = -0.44, R) >droGri2.scaffold_15203 6046604 86 + 11997470 ------------UACUU-UACUUAAACUGCCGCU-----GCGAGUGGCGCAAAUUUUACGCGACAUAAUCGUAAUAUGAUUAUC----GUGCCACAUAAUCUACACUC--- ------------.....-..........(((((.-----....)))))..........(((((..(((((((...)))))))))----))).................--- ( -16.50, z-score = -1.00, R) >consensus AACUUCCACUUAAACCU_CGCUCAAUCACUCAUUCAUACGGCAGUGGCGCAAAUUUUACGCGACGUAAUCGUAAUAUGAUUAUC____GUGCCACAUAAUC__________ .......................................((((((.(((.........))).))((((((((...))))))))......)))).................. (-10.89 = -11.18 + 0.28)

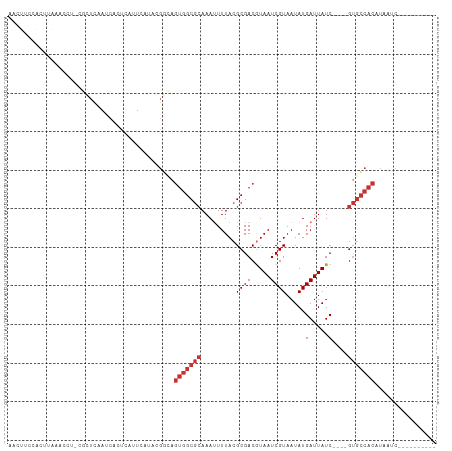
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:19 2011