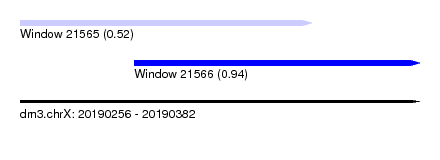
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,190,256 – 20,190,382 |
| Length | 126 |
| Max. P | 0.942239 |

| Location | 20,190,256 – 20,190,348 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 66.61 |
| Shannon entropy | 0.50688 |
| G+C content | 0.36924 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -6.68 |
| Energy contribution | -9.37 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20190256 92 + 22422827 ----ACAGAAUGCCACAGAAGUGCAUGU--GGUAUUGAAAUCACAAGAACC-UAAAAGUAUGCAGUGAAAAUACAAUUUAAAAUGUAACGCA-GCAUACU-------- ----....(((((((((........)))--))))))...............-....(((((((.(((....((((........)))).))).-)))))))-------- ( -22.40, z-score = -3.04, R) >droSec1.super_8 2375838 94 + 3762037 ----ACAGAAUGCCACAGAAGUGCAUGUGUGGUGUUGAAAUCACAAGAACC-UGAAAGUAUGCAAUGAAAAUACAUUCUAAAAUGUAGCGCA-GCAUACU-------- ----.(((.((((.((....)))))).((((((......)))))).....)-))..(((((((..((....((((((....)))))).))..-)))))))-------- ( -23.90, z-score = -1.88, R) >droSim1.chrX 15471324 94 + 17042790 ----ACAGAAUGCCACAGAAGUGCAUGUGUGGUGUUGAAAUCACAAGAACC-UGAAAGUAUGCAAUGAAAAUACAUUCUAAAAUGUAACGCA-GCAUACU-------- ----.(((.((((.((....)))))).((((((......)))))).....)-))..(((((((..((....((((((....))))))...))-)))))))-------- ( -22.60, z-score = -1.81, R) >droVir3.scaffold_13042 4532434 107 - 5191987 AGCUAACAAACGCAAAACAUAUGCAGAUG-GAUAUUCAAUUGAUGCAACCCACAAACAUCCUUUUUGCCCUUUCACUGACUAAUUUAGUGGAAGGACACGCAUUAAAU .((........((((((...(((....((-(....((....))......)))....)))...))))))(((((((((((.....)))))))))))....))....... ( -22.60, z-score = -2.64, R) >consensus ____ACAGAAUGCCACAGAAGUGCAUGUG_GGUAUUGAAAUCACAAGAACC_UAAAAGUAUGCAAUGAAAAUACAUUCUAAAAUGUAACGCA_GCAUACU________ ........(((((((((.(......).)))))))))....................(((((((........((((((....))))))......)))))))........ ( -6.68 = -9.37 + 2.69)


| Location | 20,190,292 – 20,190,382 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 61.95 |
| Shannon entropy | 0.57047 |
| G+C content | 0.33565 |
| Mean single sequence MFE | -20.91 |
| Consensus MFE | -4.94 |
| Energy contribution | -6.38 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20190292 90 + 22422827 ACAAGAACCUAAAAGUAUGCAGU--GAAAAUACAAUUUAAAAUGUAACGCAGCAUACUUUUACACACCUUUAUACAUACAU---GUGUGCUAUAU------------- .........(((((((((((.((--(....((((........)))).))).)))))))))))(((((..............---)))))......------------- ( -18.84, z-score = -2.58, R) >droSec1.super_8 2375876 86 + 3762037 ACAAGAACCUGAAAGUAUGCAAU--GAAAAUACAUUCUAAAAUGUAGCGCAGCAUACUUUUACACACCCUUAUACAU-------GUGUGAUAUGU------------- .........(((((((((((..(--(....((((((....)))))).))..)))))))))))(((((..........-------)))))......------------- ( -20.90, z-score = -3.29, R) >droSim1.chrX 15471362 90 + 17042790 ACAAGAACCUGAAAGUAUGCAAU--GAAAAUACAUUCUAAAAUGUAACGCAGCAUACUUUUACACACCCUUAUACAUACAU---GUGUGAUAUGU------------- .........(((((((((((..(--(....((((((....))))))...)))))))))))))(((((..............---)))))......------------- ( -19.24, z-score = -2.85, R) >droVir3.scaffold_13042 4532474 107 - 5191987 GAUGCAACCCACAAACAUCCUUUUUGCCCUUUCACUGACUAAUUUAGUGGAA-GGACACGCAUUAAAUUUAACACAUUCAAUUGAUGCGAUUUGCUGACCUAUUUCGU ...((((.................((.(((((((((((.....)))))))))-)).))((((((((...............))))))))..))))............. ( -24.66, z-score = -3.81, R) >consensus ACAAGAACCUAAAAGUAUGCAAU__GAAAAUACAUUCUAAAAUGUAACGCAGCAUACUUUUACACACCCUUAUACAUACAU___GUGUGAUAUGU_____________ .........(((((((((((...............................)))))))))))..........(((((.......)))))................... ( -4.94 = -6.38 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:17 2011