| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,102,916 – 12,103,050 |
| Length | 134 |
| Max. P | 0.554806 |

| Location | 12,102,916 – 12,103,018 |
|---|---|
| Length | 102 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.81 |
| Shannon entropy | 0.58362 |
| G+C content | 0.60682 |
| Mean single sequence MFE | -44.94 |
| Consensus MFE | -8.36 |
| Energy contribution | -8.12 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.85 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
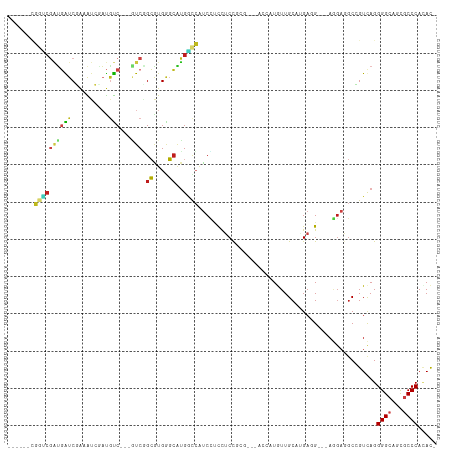
>dm3.chr2L 12102916 102 + 23011544 ------CGGUCGAUGAUCCAAAUGGAUGUC---GUCGGCGUGGGCAUGGCCAUCCUCCUCCGCGACGACCAUGUUGCAGGAGG---AGGAGGCGGUUAGGGGCAACGCCCACAC- ------..(((((((((((....)))..))---))))))((((((((.(((.((((((((((((((......))))).)))))---))))))).))....(....)))))))..- ( -59.70, z-score = -5.84, R) >droSim1.chr2L 11900560 102 + 22036055 ------CGGUCGAUGAUCCAAAUGGAUGUC---GUCGGCGUGGGCAUGGCCAUCCUCCUCCGCGACGACCAUGUUGCAGGAGG---AGGAGGCGGUUAGGGGCAACGCCCACAC- ------..(((((((((((....)))..))---))))))((((((((.(((.((((((((((((((......))))).)))))---))))))).))....(....)))))))..- ( -59.70, z-score = -5.84, R) >droSec1.super_16 291141 102 + 1878335 ------CGGUCGAUGAUCCAAAUGGAUGUC---GUCGGCGUGGGCAUGGCCAUCCUCCUCCGCGACGACCAUGUUGCAGGAGG---AGGAGGCGGUUAGGGGCAACGCCCACAC- ------..(((((((((((....)))..))---))))))((((((((.(((.((((((((((((((......))))).)))))---))))))).))....(....)))))))..- ( -59.70, z-score = -5.84, R) >droYak2.chr2L 8520308 102 + 22324452 ------UGGUCGGUGAUCCAAAUGGAUGUC---GUCGGCGUGGGCAUGGCCAUCCUCCUCCGCGACGACCAUGUUGCAGGAGG---AGGAGGCGGUUAGGGGCAACGCCCACAC- ------..((((..(((((....)))..))---..))))((((((((.(((.((((((((((((((......))))).)))))---))))))).))....(....)))))))..- ( -56.90, z-score = -4.84, R) >droEre2.scaffold_4929 13330372 102 - 26641161 ------UGGUCGGUGAUCCAAAUGGAUGUC---GUCGGCGUGGGCAUGGCCAUCCUCCUCCGCGACGACCAUGUUGCAGGAGG---AGGAGGCGGUUAGGGGCAACGCCCACAC- ------..((((..(((((....)))..))---..))))((((((((.(((.((((((((((((((......))))).)))))---))))))).))....(....)))))))..- ( -56.90, z-score = -4.84, R) >droAna3.scaffold_12943 2507840 102 - 5039921 ------CGGUCGAUGGUCGAGAUGAAUCUC---AUCCGCGUGGGCAUGGCCAUCCUCCUCUGCCACUACCAUGUUGCAUGAGG---AGGAGGCUGUCAGGGGCAGCGCCCAUAC- ------.....(.(((..((((....))))---..))))((((((.......(((((((((((.((......)).))).))))---)))).(((((.....)))))))))))..- ( -46.40, z-score = -2.30, R) >dp4.chr4_group3 911660 96 + 11692001 ------CGGGCGAUGGUCGAUAUCUAUGUC---CUCGGCAUGGGCAUGCUCCUCCGCCACC------ACCAUAUUGCAUGAUG---AGGAUGCCGUCAGGGGAAGCGCCCACAC- ------.(((((((((.......)))).((---((((((...(((((.(((.((.((....------........))..)).)---)).)))))))).)))))..)))))....- ( -32.70, z-score = 0.41, R) >droPer1.super_1 2391411 96 + 10282868 ------CGGGCGAUGGUCGAUAUCUAUGUC---CUCGGCAUGGGCAUGCUCCUCCGCCACC------ACCAUAUUGCAUGAUG---AGGAUGCCGUCAGGGGAAGCGCCCACAC- ------.(((((((((.......)))).((---((((((...(((((.(((.((.((....------........))..)).)---)).)))))))).)))))..)))))....- ( -32.70, z-score = 0.41, R) >droWil1.scaffold_181038 47012 95 + 637489 -------------CCAUCGAGAGCCCCGUC---AUCUGGGUGAUCAUGAUGACCAUCCUCUUCG---ACCAUAUUGCAAGAUGCAGACGAGGCUGUCAGGGGCAGCGCCCAUAU- -------------.....((.((((.((((---....(((((.((.....)).)))))......---.......(((.....))))))).)))).))..((((...))))....- ( -32.80, z-score = -1.04, R) >droVir3.scaffold_12963 15929650 96 + 20206255 ------CGGACGAUGAUCGAUGUCAAUGUCAAUGUCCGCAUGGGCGUGACCAUCCUCCUCC---------AUGUUGCAUGACG---AGGAGGCCGUCAGGGGCAGCGCCCAAAC- ------((((((.((((..........)))).))))))..(((((((..((..((((((((---------(((...))))..)---))))))((....))))..)))))))...- ( -41.30, z-score = -2.39, R) >droMoj3.scaffold_6500 2910690 96 + 32352404 ------GGCUCGAUGAUCAAUGUCGAUGUCAAUGUCAGCAUGAGCAUGACCGUCUUCCUCC---------AUGUUGCAUGACG---AAGAGGCCGUCAGGGGCAGCGCCCAUAC- ------(((..((((.(((.(((..((((........))))..)))))).)))).......---------..(((((.(((((---.......)))))...)))))))).....- ( -28.80, z-score = 0.36, R) >droGri2.scaffold_15252 13357862 96 + 17193109 ------GGGCCGAUGAUCGAUGUCGAUAUCGAUGUCGGCAUGUGCAUGGCCAUCUUCUUCC---------AUGUUGCAAGACG---AGGAGGCCGUUAGGGGCAGCGCCCAAAC- ------((((((((.(((((((....)))))))))))))..((((...(((.....(((((---------.((((....))))---.)))))((....))))).))))))....- ( -38.50, z-score = -1.84, R) >anoGam1.chr3R 21441117 105 + 53272125 CUAUCCUUCUCCUGCGUCGGCGUCUUCGCCU-CCUCCGGAUGGUGUGGAUGAUCACUCUCCUCGGUCACCAUGUUGCAUGAUG---------CCGUCAGGGGCAGUGCCCACACC .........(((.(.(..((((....)))).-.).).))).(((((((.....((((((((((((.((.((((...)))).))---------)))..))))).)))).))))))) ( -38.10, z-score = -1.41, R) >consensus ______CGGUCGAUGAUCGAAAUCGAUGUC___GUCGGCGUGGGCAUGGCCAUCCUCCUCCGCG___ACCAUGUUGCAUGAGG___AGGAGGCCGUCAGGGGCAGCGCCCACAC_ .......((((.(((.((.......................)).)))))))................................................((((...))))..... ( -8.36 = -8.12 + -0.24)


| Location | 12,102,946 – 12,103,050 |
|---|---|
| Length | 104 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.58 |
| Shannon entropy | 0.52644 |
| G+C content | 0.64917 |
| Mean single sequence MFE | -49.15 |
| Consensus MFE | -13.86 |
| Energy contribution | -14.15 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
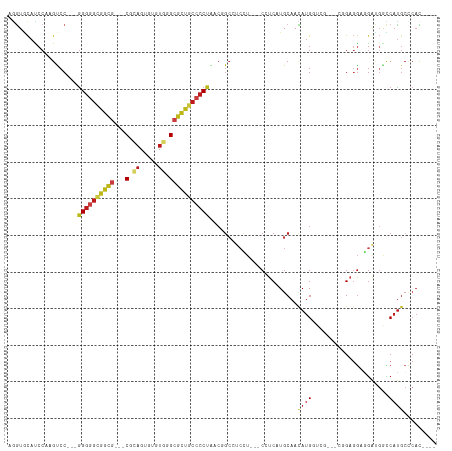
>dm3.chr2L 12102946 104 - 23011544 AGGUGCAUCCAAGUCC---GGGGGCGGCG---CGCAGUGUGUGGGCGUUGCCCCUAACCGCCUCCU---CCUCCUGCAACAUGGUCGUCGCGGAGGAGGAUGGCCAUGCCCAC---- .((.((((........---((((((((((---(.((.....)).)))))))))))....(((((((---(((((.((.((......)).))))))))))).))).))))))..---- ( -57.70, z-score = -3.31, R) >droSim1.chr2L 11900590 104 - 22036055 AGGUGCAUCCAAGUCC---GGGGGCGGCG---CGCAGUGUGUGGGCGUUGCCCCUAACCGCCUCCU---CCUCCUGCAACAUGGUCGUCGCGGAGGAGGAUGGCCAUGCCCAC---- .((.((((........---((((((((((---(.((.....)).)))))))))))....(((((((---(((((.((.((......)).))))))))))).))).))))))..---- ( -57.70, z-score = -3.31, R) >droSec1.super_16 291171 104 - 1878335 AGGUGCAUCCAAGUCC---GGGGGCGGCG---CGCAGUGUGUGGGCGUUGCCCCUAACCGCCUCCU---CCUCCUGCAACAUGGUCGUCGCGGAGGAGGAUGGCCAUGCCCAC---- .((.((((........---((((((((((---(.((.....)).)))))))))))....(((((((---(((((.((.((......)).))))))))))).))).))))))..---- ( -57.70, z-score = -3.31, R) >droYak2.chr2L 8520338 104 - 22324452 AGGUGCAUCCAAGUCC---GGGGGCGGCG---CGCAGUGUGUGGGCGUUGCCCCUAACCGCCUCCU---CCUCCUGCAACAUGGUCGUCGCGGAGGAGGAUGGCCAUGCCCAC---- .((.((((........---((((((((((---(.((.....)).)))))))))))....(((((((---(((((.((.((......)).))))))))))).))).))))))..---- ( -57.70, z-score = -3.31, R) >droEre2.scaffold_4929 13330402 104 + 26641161 AGGUGCAUCCAAGUCC---GGGGGCGGCG---CGUAGUGUGUGGGCGUUGCCCCUAACCGCCUCCU---CCUCCUGCAACAUGGUCGUCGCGGAGGAGGAUGGCCAUGCCCAC---- .((.((((........---((((((((((---(.((.....)).)))))))))))....(((((((---(((((.((.((......)).))))))))))).))).))))))..---- ( -55.90, z-score = -3.13, R) >droAna3.scaffold_12943 2507870 104 + 5039921 AGGUGCACCCGAGUCC---GGGGGCAGCG---CGCAGUGUAUGGGCGCUGCCCCUGACAGCCUCCU---CCUCAUGCAACAUGGUAGUGGCAGAGGAGGAUGGCCAUGCCCAC---- .((.(((.....(((.---((((((((((---(.((.....)).)))))))))))))).(((((((---((((.(((.((......)).))))))))))).)))..)))))..---- ( -63.10, z-score = -4.89, R) >dp4.chr4_group3 911690 98 - 11692001 AGAUUCACCCCAGCCC---GGGGGCGGCG---CGGAGUGUGUGGGCGCUUCCCCUGACGGCAUCCU---CAUCAUGCAAUAUGGU------GGUGGCGGAGGAGCAUGCCCAU---- .(.(((.(((((((((---(((((.((((---(...........))))).))))))..)))....(---(((((((...))))))------))))).)).))).)........---- ( -36.40, z-score = 1.68, R) >droPer1.super_1 2391441 98 - 10282868 AGAUUCACCCCAGCCC---GGGGGCGGCG---CGGAGUGUGUGGGCGCUUCCCCUGACGGCAUCCU---CAUCAUGCAAUAUGGU------GGUGGCGGAGGAGCAUGCCCAU---- .(.(((.(((((((((---(((((.((((---(...........))))).))))))..)))....(---(((((((...))))))------))))).)).))).)........---- ( -36.40, z-score = 1.68, R) >droWil1.scaffold_181038 47035 110 - 637489 AAGUGCAUCCUAGUCCCGGGGGGGCGGCGGCCCGUAGCAUAUGGGCGCUGCCCCUGACAGCCUCGUCUGCAUCUUGCAAUAUGGUCG---AAGAGGAUGGUCAUCAUGAUCAC---- ..(((((((((..(((((.((((((((((.((((((...))))))))))))))))(((......)))(((.....)))...)))..)---)..))))))(((.....))))))---- ( -45.70, z-score = -1.17, R) >droVir3.scaffold_12963 15929683 95 - 20206255 AGGUACACCCCAGUCC---GGGGGCAGCG---CGCAGCGUUUGGGCGCUGCCCCUGACGGCCUCCU---CGUCAUGCAACAUG---------GAGGAGGAUGGUCACGCCCAU---- .(((........(((.---((((((((((---(.(((...))).))))))))))))))((((((((---(.(((((...))))---------)..))))).))))..)))...---- ( -51.10, z-score = -3.15, R) >droMoj3.scaffold_6500 2910723 95 - 32352404 AGGUACACCCCAGCCC---AGGGGCAGCG---CGCAGUGUAUGGGCGCUGCCCCUGACGGCCUCUU---CGUCAUGCAACAUG---------GAGGAAGACGGUCAUGCUCAU---- .((......))(((.(---((((((((((---(.((.....)).))))))))))))..((((((((---(.(((((...))))---------)..))))).))))..)))...---- ( -48.90, z-score = -3.53, R) >droGri2.scaffold_15252 13357895 95 - 17193109 AGGUACACCCCAGUCC---GGGGGCAGCG---CGCAGUGUUUGGGCGCUGCCCCUAACGGCCUCCU---CGUCUUGCAACAUG---------GAAGAAGAUGGCCAUGCACAU---- .((.((......))))---((((((((((---(.(((...))).)))))))))))...((((...(---(.((((.(.....)---------.)))).)).))))........---- ( -42.80, z-score = -2.08, R) >anoGam1.chr3R 21441154 100 - 53272125 --GUGCAUCCGAGUCCU--GGUAGUGCUCGCACUUUG-GUGUGGGCACUGCCCCUGACGG---------CAUCAUGCAACAUGGUGA---CCGAGGAGAGUGAUCAUCCACACCAUC --(((..(((..(((..--((((((((((((((....-))))))))))))))...)))((---------(((((((...))))))).---))..)))((....))...)))...... ( -45.30, z-score = -2.93, R) >triCas2.ChLG4 1461532 92 + 13894384 AAGUGUUUCCAAGUCC---GGGCACUGCC---CGAAGCGUGUGGGCUUUGCCCCAAA------------CCUCCUGCAACAUGGUCG---CAGACACCGACCAUCCUGCCACC---- ..(((...........---(((((..(((---((.......)))))..)))))....------------......(((..(((((((---.......)))))))..)))))).---- ( -31.70, z-score = -1.70, R) >consensus AGGUGCAUCCAAGUCC___GGGGGCGGCG___CGCAGUGUGUGGGCGCUGCCCCUAACGGCCUCCU___CCUCAUGCAACAUGGUCG___CGGAGGAGGAUGGCCAUGCCCAC____ ...................((((((((((...((.......))..)))))))))).............................................................. (-13.86 = -14.15 + 0.29)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:25 2011