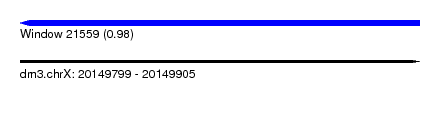
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,149,799 – 20,149,905 |
| Length | 106 |
| Max. P | 0.976263 |

| Location | 20,149,799 – 20,149,905 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 66.64 |
| Shannon entropy | 0.64234 |
| G+C content | 0.46249 |
| Mean single sequence MFE | -31.71 |
| Consensus MFE | -12.47 |
| Energy contribution | -13.08 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20149799 106 - 22422827 UGUGCUCC-GAAUGACUGCGGAUAUCCGCACUGCCGGUAAAUGUCUGCAAAGAAACU------------UCCAGUGCGGAUAUUCA-AGCUGUGCAGACUAGACGGCUACUAAUUAUAAA- ...((...-........))((((((((((((((..(((.....(((....))).)))------------..)))))))))))))).-((((((.(......)))))))............- ( -33.60, z-score = -2.80, R) >droPer1.super_13 1442640 120 - 2293547 UGUGCUCCGGAAUGACUACGGAUAUCCACAGUGGAGGAUAAUGACUGUCAAGCAGCUAAGGAUACGGCAUCUAAUGCGAAUAUUCA-GUCAGCUACAAAUUCGAAUACACUUGAUUAUAAA (((((((((.........))))(((((........))))).((((((.......((...((((.....))))...)).......))-)))))).)))...((((......))))....... ( -24.24, z-score = 0.23, R) >dp4.chrXL_group1e 5411750 120 + 12523060 UGUGCUCCGGAAUGACUACGGAUAUCCACAGUGGAGGAUAAUGACUGUCAAGCAGCUAAGGAUACGGCAUCUAAUGCGAAUAUUCA-GUCAGCUACAAAUUCGAAUACACUUGAUCAUAAA (((((((((.........))))(((((........))))).((((((.......((...((((.....))))...)).......))-)))))).)))...((((......))))....... ( -24.24, z-score = 0.39, R) >droAna3.scaffold_13417 5349584 84 + 6960332 UAUUCUGC-GAAUAGCUGCGGAUAUCCGCUUUAGUGGUAUUAG----------AGCU------------GCAAGAGCGGAUAUUCACUGCCGGGACGGAUAUUUGGG-------------- .......(-(((((.....(((((((((((((.(..((.....----------.)).------------.).)))))))))))))....(((...))).))))))..-------------- ( -29.50, z-score = -2.56, R) >droEre2.scaffold_4690 10242088 106 - 18748788 UGUGCUCC-GAAUGACUGCGGAUAUCCGCACUGCCGGUAAAUGUCCGCAAAGCAACU------------UCCUGCGCGGAUAUUCA-AGCUGUGCAGACUAGACGCGUACUAAUUAUAAA- .((((..(-(......(((((....)))))((((((((.(((((((((...(((...------------...))))))))))))..-.)))).))))......)).))))..........- ( -36.10, z-score = -2.95, R) >droYak2.chrX 19388640 106 - 21770863 UGUGCUCC-GAAUGACUGCGGAUAUCCGCACUGCCGGUAAAUGUCUGCAAGGAAACU------------UCCAGUGCGGAUAUUCA-AGCUG-UGCAGACUAGACGAGUACUAAUUAUAAA .((((((.-.......(((((....)))))((((((((.((((((((((.(((....------------)))..))))))))))..-.))))-.)))).......)))))).......... ( -40.10, z-score = -4.68, R) >droSec1.super_8 2343974 106 - 3762037 UGUGCUCC-GAAUGACUGCGGAUAUCCGCACUGCCGGUAAUUGUCUGCAAAGAAACU------------UCCCGUGCGGAUAUUCA-AGCUGUGCCGACUAGACGGGUACUAAUUAUCAA- .((((.((-(......(((((....)))))(.((((((...((((((((..((....------------))...))))))))....-.)))).)).)......)))))))..........- ( -32.00, z-score = -1.79, R) >droSim1.chrX 15456038 106 - 17042790 UGUGCUCC-GAAUGACUGCGGAUAUCCGCACUGCCGGUAAAUGUCUGCAAAGAAACU------------UCCCGUGCGGAUAUUCA-AGCUGUGCCGACUAGACGGGUACUAAUUAUAAA- .((((.((-(......(((((....)))))(.((((((.((((((((((..((....------------))...))))))))))..-.)))).)).)......)))))))..........- ( -33.90, z-score = -2.78, R) >consensus UGUGCUCC_GAAUGACUGCGGAUAUCCGCACUGCCGGUAAAUGUCUGCAAAGAAACU____________UCCAGUGCGGAUAUUCA_AGCUGUGCCGAAUAGACGGGUACUAAUUAUAAA_ ...................((((((((((((..........................................)))))))))))).................................... (-12.47 = -13.08 + 0.61)

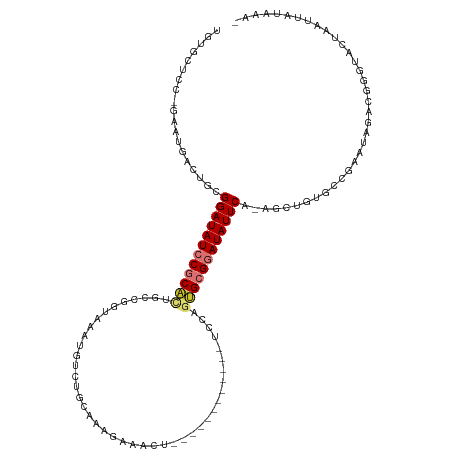
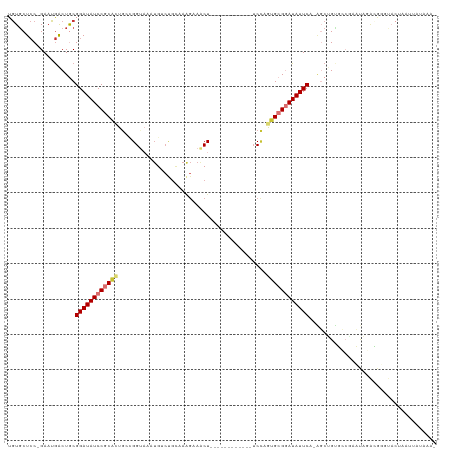
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:11 2011