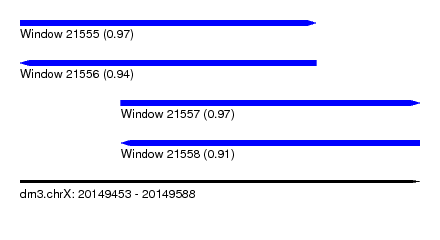
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,149,453 – 20,149,588 |
| Length | 135 |
| Max. P | 0.970875 |

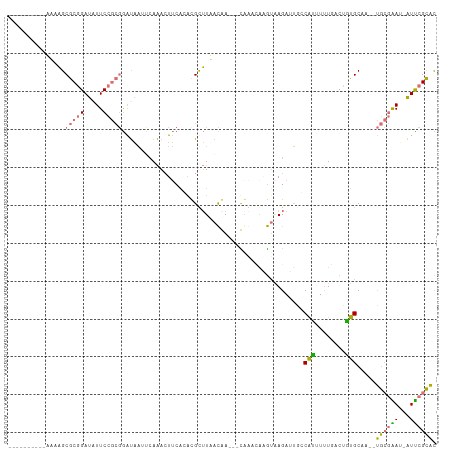
| Location | 20,149,453 – 20,149,553 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 52.02 |
| Shannon entropy | 0.75495 |
| G+C content | 0.44817 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -10.93 |
| Energy contribution | -11.18 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20149453 100 + 22422827 GUGCGAAUUGUCCGCAUAUUGCACAGGCAAAAAUGCUAAUCUUACUUGGCCG---UCGUAAAACGCGUGAAGUUUGACUUAUCCGCGGAAUAUCCGCGCUUUU---------- (((((.....(((((...((((....)))).........((..((((.((((---(......))).)).))))..)).......))))).....)))))....---------- ( -29.90, z-score = -1.73, R) >droEre2.scaffold_4690 10242042 106 + 18748788 ACACUAAC-UUUUCUGCCGUGCAAGCUUAGUUUUGGCGUUCUU-UUUGUUUAUAAUUAGUACGCGUCU--AGUCUGCACAGCUU---GAAUAUCCGCGCAGGAAGUUGCUUUG ....((((-(((.((((.(((((..((.((.....((((.((.-.(((....)))..)).))))..))--))..))))).((..---........)))))))))))))..... ( -27.00, z-score = -0.88, R) >droYak2.chrX 19388325 99 + 21770863 GUGCGAAU-AUUCGCA----GCGUAGCAAAACACGGCACUCUCACGCCUCUAAUUUUGUUAAGCAUUCGAAUUUUGA-UUAUCCGCGGAAUAUCCGCGCUGAAAG-------- (((((.((-((((((.----((.((((((((...(((........))).....)))))))).))..((((...))))-......)).)))))).)))))......-------- ( -27.20, z-score = -1.72, R) >droSim1.chrX 15455696 97 + 17042790 GUGCGGAU-AUCCGCA--UUGCACAGUCAAAAAUGCGAAUCUUACCUAGUCG---UCGUAAAACGUGUGAAGUUUUGAUUAUCCGCGGAACAUCCGCGCUUUU---------- ((((((((-.(((((.--(..(((.((......(((((..((.....))...---)))))..)))))..)......(......))))))..))))))))....---------- ( -29.00, z-score = -2.21, R) >consensus GUGCGAAU_AUCCGCA__UUGCACAGUCAAAAAUGCCAAUCUUACUUGGCCA___UCGUAAAACGUGUGAAGUUUGAAUUAUCCGCGGAAUAUCCGCGCUGUA__________ .(((((.....)))))....((....................................((((((.......)))))).......((((.....)))))).............. (-10.93 = -11.18 + 0.25)

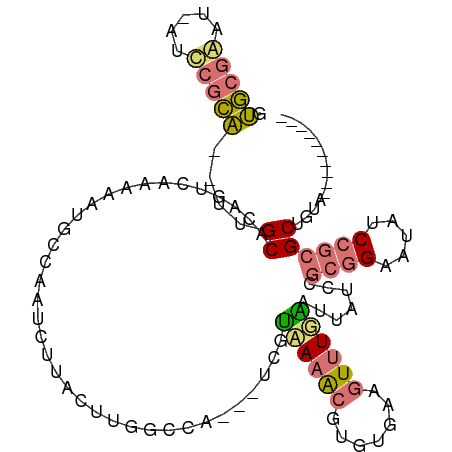
| Location | 20,149,453 – 20,149,553 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 52.02 |
| Shannon entropy | 0.75495 |
| G+C content | 0.44817 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -10.94 |
| Energy contribution | -10.88 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.937765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20149453 100 - 22422827 ----------AAAAGCGCGGAUAUUCCGCGGAUAAGUCAAACUUCACGCGUUUUACGA---CGGCCAAGUAAGAUUAGCAUUUUUGCCUGUGCAAUAUGCGGACAAUUCGCAC ----------......((((((..((((((....((((..((((..((((.....)).---))...))))..)))).((((........))))....))))))..)))))).. ( -28.40, z-score = -1.53, R) >droEre2.scaffold_4690 10242042 106 - 18748788 CAAAGCAACUUCCUGCGCGGAUAUUC---AAGCUGUGCAGACU--AGACGCGUACUAAUUAUAAACAAA-AAGAACGCCAAAACUAAGCUUGCACGGCAGAAAA-GUUAGUGU ....(((((((((.....))).....---..((((((((..((--((..((((.((.............-.)).)))).....)).))..)))))))).....)-))).)).. ( -22.44, z-score = -0.04, R) >droYak2.chrX 19388325 99 - 21770863 --------CUUUCAGCGCGGAUAUUCCGCGGAUAA-UCAAAAUUCGAAUGCUUAACAAAAUUAGAGGCGUGAGAGUGCCGUGUUUUGCUACGC----UGCGAAU-AUUCGCAC --------......((((((.....))))(((((.-((...((((..((((((...........))))))..))))((.((((......))))----.)))).)-)))))).. ( -29.60, z-score = -1.46, R) >droSim1.chrX 15455696 97 - 17042790 ----------AAAAGCGCGGAUGUUCCGCGGAUAAUCAAAACUUCACACGUUUUACGA---CGACUAGGUAAGAUUCGCAUUUUUGACUGUGCAA--UGCGGAU-AUCCGCAC ----------......((((((((.(((((...((((...((((....((((....))---))...))))..)))).((((........))))..--)))))))-)))))).. ( -29.30, z-score = -2.42, R) >consensus __________AAAAGCGCGGAUAUUCCGCGGAUAAUUCAAACUUCACACGCUUAACAA___CAAACAAGUAAGAUUGCCAUUUUUGACUGUGCAA__UGCGAAU_AUUCGCAC ...............(((((.....)))))...............................................((((........))))....((((((...)))))). (-10.94 = -10.88 + -0.06)

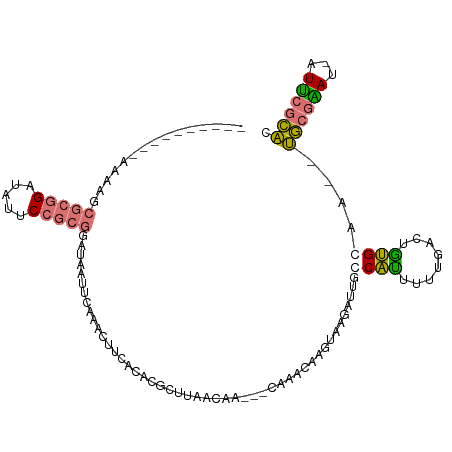
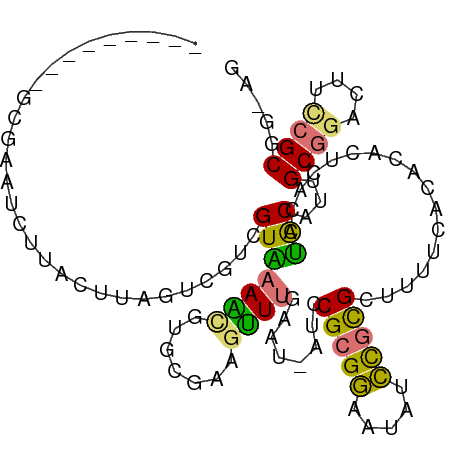
| Location | 20,149,487 – 20,149,588 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 53.69 |
| Shannon entropy | 0.76602 |
| G+C content | 0.49234 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -12.56 |
| Energy contribution | -11.50 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20149487 101 + 22422827 ---------GCUAAUCUUACUUGGCCGUCGUAAAACGCGUGAAGUUUGACUUAUCCGCGGAAUAUCCGCGCUUUUCACACACUCGCCUACAUUGCGGACUUUCGCGGGAG ---------.....((..((((.(((((......))).)).))))..))....(((((((((..((((((......................)))))).))))))))).. ( -28.45, z-score = -1.21, R) >droEre2.scaffold_4690 10242075 108 + 18748788 GGCGUUCUUUUUGUUUAUAAUUAGUACGCGUCUAGUCUGCACAGCUUGAAU-AUCCGCGCAGGAAGU-UGCUUUGCGGACAUUUACCGGCAGUGCGGAUAUCCGCAGUCA (((((.((..(((....)))..)).))(((..((.(((((((.((((((((-.(((((((((....)-)))...))))).)))))..))).)))))))))..))).))). ( -35.70, z-score = -1.97, R) >droYak2.chrX 19388354 94 + 21770863 GGCACUCUCACGCCUCUAAUUU------UGUUAAGCAUUCGAAUUUUGAUU-AUCCGCGGAAUAUCCGCGCU----GAAAGAUCAGCAGCA--GCCAUCUACUGCGA--- (((........)))........------.......................-....((((.....))))(((----((....))))).(((--(.......))))..--- ( -22.80, z-score = -0.61, R) >droSim1.chrX 15455727 101 + 17042790 ---------GCGAAUCUUACCUAGUCGUCGUAAAACGUGUGAAGUUUUGAUUAUCCGCGGAACAUCCGCGCUUUUUACACACUCGCCUACAUUGCGGACUUCCGCGGCAG ---------((((..((.....))...)))).....((((((((...........(((((.....)))))..))))))))....(((......((((....))))))).. ( -28.02, z-score = -1.37, R) >consensus _________GCGAAUCUUACUUAGUCGUCGUAAAACGUGCGAAGUUUGAAU_AUCCGCGGAAUAUCCGCGCUUUUCACACACUCACCUACAUUGCGGACUUCCGCGG_AG .............................(((((((.......))))........(((((.....))))).................)))...((((....))))..... (-12.56 = -11.50 + -1.06)

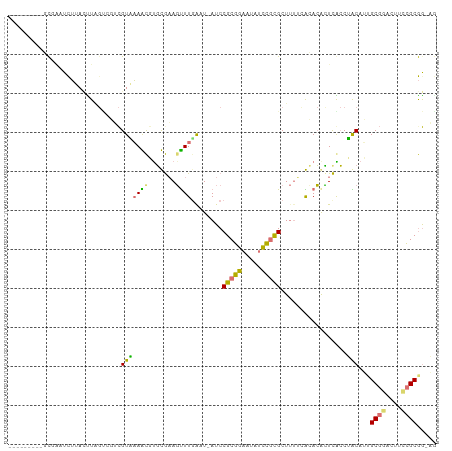
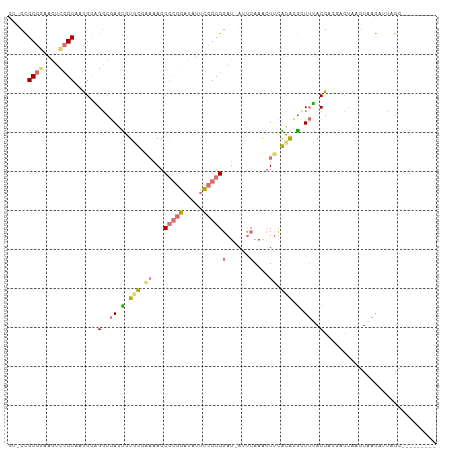
| Location | 20,149,487 – 20,149,588 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 53.69 |
| Shannon entropy | 0.76602 |
| G+C content | 0.49234 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -10.74 |
| Energy contribution | -13.30 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20149487 101 - 22422827 CUCCCGCGAAAGUCCGCAAUGUAGGCGAGUGUGUGAAAAGCGCGGAUAUUCCGCGGAUAAGUCAAACUUCACGCGUUUUACGACGGCCAAGUAAGAUUAGC--------- ((..((((......)))......(((....(((((((...(((((.....)))))((....))....)))))))(((....))).)))..)..))......--------- ( -27.30, z-score = -0.04, R) >droEre2.scaffold_4690 10242075 108 - 18748788 UGACUGCGGAUAUCCGCACUGCCGGUAAAUGUCCGCAAAGCA-ACUUCCUGCGCGGAU-AUUCAAGCUGUGCAGACUAGACGCGUACUAAUUAUAAACAAAAAGAACGCC ....(((((....)))))((((((((.(((((((((...(((-......)))))))))-)))...)))).)))).......((((.((..............)).)))). ( -35.44, z-score = -3.52, R) >droYak2.chrX 19388354 94 - 21770863 ---UCGCAGUAGAUGGC--UGCUGCUGAUCUUUC----AGCGCGGAUAUUCCGCGGAU-AAUCAAAAUUCGAAUGCUUAACA------AAAUUAGAGGCGUGAGAGUGCC ---..(((((.....))--))).(((((....))----)))((((.....))))....-.......((((..((((((....------.......))))))..))))... ( -29.20, z-score = -1.44, R) >droSim1.chrX 15455727 101 - 17042790 CUGCCGCGGAAGUCCGCAAUGUAGGCGAGUGUGUAAAAAGCGCGGAUGUUCCGCGGAUAAUCAAAACUUCACACGUUUUACGACGACUAGGUAAGAUUCGC--------- ((((.((((....))))...))))(((((((((.......(((((.....)))))(.....).......)))))(((((((.........)))))))))))--------- ( -31.00, z-score = -1.04, R) >consensus CU_CCGCGGAAGUCCGCAAUGCAGGCGAGUGUGCGAAAAGCGCGGAUAUUCCGCGGAU_AUUCAAACUUCACACGCUUUACGACGACUAAGUAAGAUUAGC_________ .....((((....))))......(.((((((((((((...(((((.....)))))((....))....)))))))))))).)............................. (-10.74 = -13.30 + 2.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:10 2011