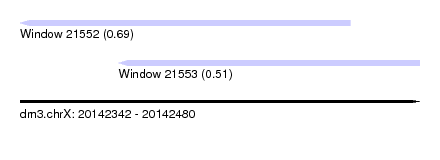
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,142,342 – 20,142,480 |
| Length | 138 |
| Max. P | 0.690778 |

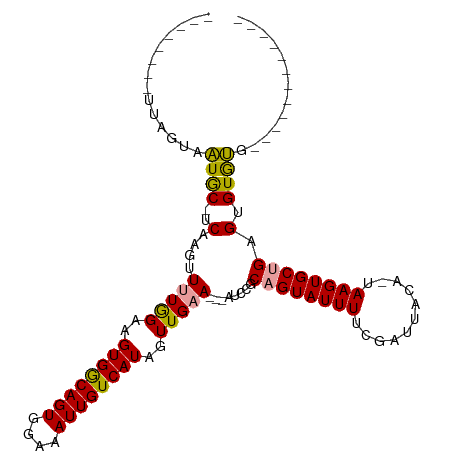
| Location | 20,142,342 – 20,142,456 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.39 |
| Shannon entropy | 0.31415 |
| G+C content | 0.38626 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -16.19 |
| Energy contribution | -15.53 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.690778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20142342 114 - 22422827 UUGUAUCUUGAGCGGUACUUCACGUUCUAGAAGUGACAGUGGAAAUUGUCAUCGUUGAAAUUAUCCGCAGUAUUUUCGAUUACAUUAAGUGCUGAGUGUACAGAAGCUGCAGUA ..(((..((.(((((((((((........)))))((((((....)))))))))))).))..)))..(((((...(((...((((((........))))))..)))))))).... ( -30.10, z-score = -1.18, R) >droSec1.super_8 2335592 89 - 3762037 --------UUAGUAAUGC-UCAAGUUUUGGAAGUGGCAGUGGAAAUUGUCAUAGUUGAA---AUCCGCAGUAUUUUCGAUUACA-UAAGUGCUGAGUGUGUG------------ --------......((((-((..((((..(..((((((((....))))))))..)..))---))....(((((((.........-.)))))))))))))...------------ ( -22.40, z-score = -1.96, R) >droSim1.chrX_random 5231802 89 - 5698898 --------UUAGUAAUGC-UCAAGUUUUGGAAGUGGCAGUGGAAAUUGUCAUAGUUGAA---AUCCGCUGUAUUUUCGAUUACA-UAAGUGCUGAGUGUGUG------------ --------......((((-(((....(((((((((.(((((((.....(((....))).---.)))))))))))))))).(((.-...))).)))))))...------------ ( -24.00, z-score = -2.55, R) >consensus ________UUAGUAAUGC_UCAAGUUUUGGAAGUGGCAGUGGAAAUUGUCAUAGUUGAA___AUCCGCAGUAUUUUCGAUUACA_UAAGUGCUGAGUGUGUG____________ ..............((((.......(((((..((((((((....))))))))..)))))........((((((((...........))))))))...))))............. (-16.19 = -15.53 + -0.66)

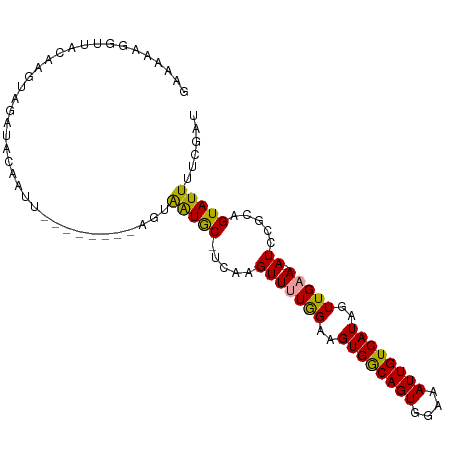
| Location | 20,142,376 – 20,142,480 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Shannon entropy | 0.22074 |
| G+C content | 0.34727 |
| Mean single sequence MFE | -22.37 |
| Consensus MFE | -13.42 |
| Energy contribution | -13.20 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.509531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20142376 104 - 22422827 UAAAAAGCUUACAAAUAGAUACAAUUGUAUCUUGAGCGGUACUUCACGUUCUAGAAGUGACAGUGGAAAUUGUCAUCGUUGAAAUUAUCCGCAGUAUUUUCGAU ......(((((.....((((((....)))))))))))((((((((........)))))((((((....)))))))))(((((((.(((.....))).))))))) ( -26.60, z-score = -2.68, R) >droSec1.super_8 2335613 92 - 3762037 GAAAAAGGUUACAAGUAGAUACAAUU--------AGUAAUGC-UCAAGUUUUGGAAGUGGCAGUGGAAAUUGUCAUAGUUGAA---AUCCGCAGUAUUUUCGAU (((((..(((((.(((.......)))--------.)))))((-((..((((..(..((((((((....))))))))..)..))---))..).))).)))))... ( -19.90, z-score = -2.11, R) >droSim1.chrX_random 5231823 92 - 5698898 GAAAAAGGUUACAAGUAGAUACAAUU--------AGUAAUGC-UCAAGUUUUGGAAGUGGCAGUGGAAAUUGUCAUAGUUGAA---AUCCGCUGUAUUUUCGAU .............((((..(((....--------.))).)))-)......(((((((((.(((((((.....(((....))).---.)))))))))))))))). ( -20.60, z-score = -1.99, R) >consensus GAAAAAGGUUACAAGUAGAUACAAUU________AGUAAUGC_UCAAGUUUUGGAAGUGGCAGUGGAAAUUGUCAUAGUUGAA___AUCCGCAGUAUUUUCGAU ................((((((..................................((((((((....)))))))).((.((.....)).)).))))))..... (-13.42 = -13.20 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:06 2011