| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,101,363 – 12,101,463 |
| Length | 100 |
| Max. P | 0.526891 |

| Location | 12,101,363 – 12,101,463 |
|---|---|
| Length | 100 |
| Sequences | 14 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.55 |
| Shannon entropy | 0.61086 |
| G+C content | 0.42795 |
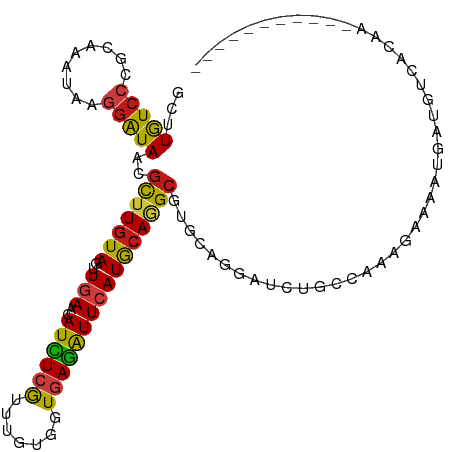
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -13.04 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.526891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
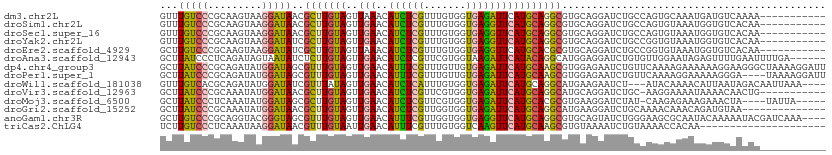
>dm3.chr2L 12101363 100 + 23011544 GUUUGUCCCGCAAGUAAGGAUAACGCUUGUAGUUAAACAUCUCGUUUGUGGUGAGAUUCAUGCAGGCGUGCAGGAUCUGCCAGUGCAAAUGAUGUCAAAA----------- (((((..(..(((((.........)))))..).)))))...(((((((((((.((((((.((((....)))))))))))))...))))))))........----------- ( -27.60, z-score = -0.97, R) >droSim1.chr2L 11899009 100 + 22036055 GUUUGUCCCGCAAGUAAGGAUAACGCUUGUAGUUGAACAUCUCGUUUGUGGUGAGGUUCAUGCAGGCGUGCAGGAUCUGCCAGUGUAAAUGGUGUCACAA----------- ...((((((....)...)))))(((((((((..(((((..((((.......))))))))))))))))))....(((..((((.......)))))))....----------- ( -28.00, z-score = -0.43, R) >droSec1.super_16 289588 100 + 1878335 GUUUGUCCCGCAAGUAAGGAUAACGCUUGUAGUUGAACAUCUCGUUUGUGGUGAGGUUCAUGCAGGCGUGCAGGAUCUGCCAGUGUAAAUGGUGUCACAA----------- ...((((((....)...)))))(((((((((..(((((..((((.......))))))))))))))))))....(((..((((.......)))))))....----------- ( -28.00, z-score = -0.43, R) >droYak2.chr2L 8518755 100 + 22324452 GUUUGUCCCGCAAGUAAGGAUAUCGCUUGUAGUUGAACAUCUCGUUUGUGGUGAGGUUCAUGCAGGCGUGCAGGAUCUGCCGGUGUAAAUGGUGUCACAA----------- ..((((..(((........((((((((((((..(((((..((((.......))))))))))))))))(.((((...)))))))))).....)))..))))----------- ( -26.32, z-score = 0.21, R) >droEre2.scaffold_4929 13328819 100 - 26641161 GCUUGUCCCGCAAGUAAGGAUAUCGCUUGUAGUUAAACAUCUCGUUUGUGGUGAGGUUCAUGCACGCGUGCAGGAUCUGCCGGUGUAAAUGGUGUCACAA----------- (((((.....)))))...(((((((((..(...(((((.....)))))..)..)).....(((((.((.((((...)))))))))))..)))))))....----------- ( -27.70, z-score = -0.11, R) >droAna3.scaffold_12943 2506287 105 - 5039921 GCUUAUCCCUCAGAUAGUAAUAUCUCUUGUAGUUGAACAUCUCGUUCGUGGUAAGAUUCAUACAGGCAUGGAGGAUCUGUGUUGGAAUAGAGUUUUGAAUUUUGA------ .......(((((((((....)))))(((((....((((.....))))((((......)))))))))....)))).(((((......)))))..............------ ( -21.20, z-score = 0.74, R) >dp4.chr4_group3 910099 111 + 11692001 GCUUAUCCCGCAGAUAUGGAUAGCGUUUGUAGUUGAACAUUUCGUUUGUUGUGAGAUUCAUGCAAGCGUGGAGAAUCUGUUCAAAAGAAAAAAGGAAGGGCUAAAAGGAUU (((..(((.((((((.......(((((((((..(((..((((((.......)))))))))))))))))).....))))))((....)).....)))..))).......... ( -25.40, z-score = -0.63, R) >droPer1.super_1 2389855 107 + 10282868 GCUUAUCCCGCAGAUAUGGAUAGCGUUUGUAGUUGAACAUUUCGUUUGUUGUGAGAUUCAUGCAAGCGUGGAGAAUCUGUUCAAAAGGAAAAAGGGA----UAAAAGGAUU ..(((((((((((((.......(((((((((..(((..((((((.......)))))))))))))))))).....))))))((.....))....))))----)))....... ( -27.50, z-score = -1.82, R) >droWil1.scaffold_181038 45505 104 + 637489 GUUUGUCACGCAGAUAUGGAUAUCGUUUAUAGUUGAACAUCUCAUUUGUGGUGAGAUUCAUGCAGGCAUGAAGAAUCU---AUACAAAACAUUAAUAGACAAUUAAA---- ..(((((.(((((((..((((...(((((....))))))))).)))))))((((((((((((....)))))...))))---.)))............))))).....---- ( -23.40, z-score = -1.51, R) >droVir3.scaffold_12963 15928102 99 + 20206255 GCUUAUCCCGCAAAUAUGGAUAACGCUUGUAGUUGAACAUCUCGUUCGUGGUGAGAUUCAUGCAGGCAUGCAGGAUCUGC-AAGGAAAAUAAAACAACUG----------- ((..((((.(((.....(.....)(((((((..(((..((((((.......)))))))))))))))).))).))))..))-...................----------- ( -25.10, z-score = -1.00, R) >droMoj3.scaffold_6500 2909144 101 + 32352404 GCUUAUCCCUCAAAUAUGGAUAGCGCUUGUAGUUGAACAUCUCGUUCGUGGUGAGAUUCAUGCACGCGUGAAGGAUCUAU-CAAGAGAAAGAAACUA----UAUUA----- ((((((((.........)))))).)).(((((((.....((((.....((((((..((((((....))))))...)).))-)).))))....)))))----))...----- ( -25.60, z-score = -1.18, R) >droGri2.scaffold_15252 13356322 97 + 17193109 GCUUAUCCCGCAAAUAUGGAUAACGCUUGUAGUUGAACAUCUCGUUGGUGGUGAGAUUCAUGCAGGCAUGAAGGAUCUGCAAAACAAACAGAUGUAA-------------- ((((((((.........)))))).))..........((((((.(((.((..(((((((((((....)))))...)))).))..)).)))))))))..-------------- ( -25.30, z-score = -1.06, R) >anoGam1.chr3R 21439475 107 + 53272125 GCUUGUCCCGCAGGUACGGGUAGCGUUUGUAGUUGAACAUUUCGUUGGUGGUGAGGUUCAUGCAGGCGUGCAGUAUCUGGGAAGCGCAAUACAAAAAUACGAUCAAA---- ((((...((.(((((((..(((.((((((((..(((((...(((....)))....)))))))))))))))).)))))))))))))......................---- ( -30.80, z-score = -0.59, R) >triCas2.ChLG4 1455614 90 - 13894384 UCUUGUCCCUCAAAUAAGGAUAACGUUUGUAAUUGAACAUUUCGUUUGUGGUCAAGUUCAUGCAAGCGUGUAAAAUCUGUAAAACCACAA--------------------- ...(((((.........)))))(((((((((..(((((...(((....)))....)))))))))))))).....................--------------------- ( -16.70, z-score = -1.17, R) >consensus GCUUGUCCCGCAAAUAAGGAUAACGCUUGUAGUUGAACAUCUCGUUUGUGGUGAGAUUCAUGCAGGCGUGCAGGAUCUGCCAAAGAAAAUGAUGUCACAA___________ ...(((((.........)))))..(((((((..(((..((((((.......))))))))))))))))............................................ (-13.04 = -12.50 + -0.54)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:24 2011