
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,075,404 – 20,075,474 |
| Length | 70 |
| Max. P | 0.523319 |

| Location | 20,075,404 – 20,075,474 |
|---|---|
| Length | 70 |
| Sequences | 6 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 47.99 |
| Shannon entropy | 1.00321 |
| G+C content | 0.58605 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -8.06 |
| Energy contribution | -7.12 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.52 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.523319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
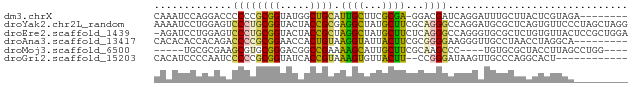
>dm3.chrX 20075404 70 - 22422827 CAAAUCCAGGACCCCCCGCGGUAUGGCUGCAUUGCUUCGCGA-GGACGAUCAGGAUUUGCUUACUCGUAGA-------- (((((((..((.(.(((((((...(((......)))))))).-))..).)).)))))))............-------- ( -19.00, z-score = 0.01, R) >droYak2.chr2L_random 3192424 79 + 4064425 AAAAUCCUGGAGUCCCUGCGGUACUACCGCGAGGCUAUGCUUCGCAGGGCCAGGAUGCGCUCAGUGUUCCCUAGCUAGG ...(((((((...((((((((.....))..(((((...))))))))))))))))))..(((.((......))))).... ( -31.80, z-score = -1.56, R) >droEre2.scaffold_1439 6821 78 - 11204 -AGAUCCUGGAGUCCCUGCGGUACUACCGCUAGGCUAUGCUUCUCAGGGCCAGGGUGCGCUCUGUGUUACUCCGCUGGA -...(((((((((((((((((.....))))..((((.((.....)).)))))))).((((...)))).))))))..))) ( -27.20, z-score = 0.39, R) >droAna3.scaffold_13417 2435071 70 + 6960332 CACACACCACAGACCCCGCGGAACCACUGUAAGGUAUUACUUCGCGGGGAAGGGUUGCCUAACCUAGGCA--------- .....(((.....((((((((((((.......))).....)))))))))...)))((((((...))))))--------- ( -25.90, z-score = -2.35, R) >droMoj3.scaffold_6500 352690 66 + 32352404 -----UGCGCGAAGCGUGCGGGACGGCCGAAAAGCAUUGCUUCGCAAGCCC----UGUGCGCUACCUUAGCCUGG---- -----.((..(.((((..((((.(...(((..(((...))))))...).))----))..)))).)....))....---- ( -20.30, z-score = 1.39, R) >droGri2.scaffold_15203 11064289 65 + 11997470 CACAUCCCCAAUCCCCCGCGGUAUCACCGUAAAGUGUUACUU--CCGGGAUAAGUUGCCCAGGCACU------------ ..............((((.((((.(((......))).)))).--.))))...(((.((....)))))------------ ( -15.30, z-score = -1.01, R) >consensus CAAAUCCCGGAGACCCCGCGGUACCACCGCAAGGCUUUGCUUCGCAGGGCCAGGAUGCGCUCACUCUUAC_________ .............((((((((.....)))).((((...))))...)))).............................. ( -8.06 = -7.12 + -0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:58 2011