| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,067,580 – 20,067,674 |
| Length | 94 |
| Max. P | 0.990769 |

| Location | 20,067,580 – 20,067,674 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.88 |
| Shannon entropy | 0.70626 |
| G+C content | 0.42814 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -13.73 |
| Energy contribution | -14.04 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
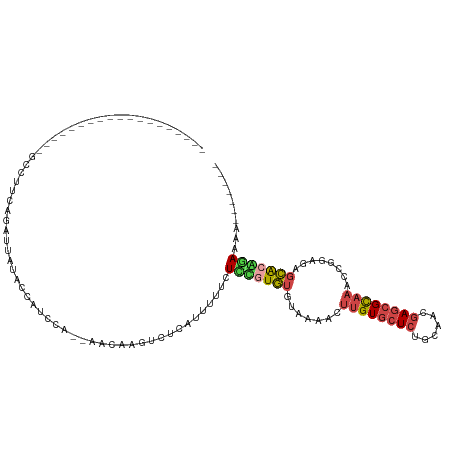
>dm3.chrX 20067580 94 - 22422827 -------------------GCCUUCAGAUUCUGCCAUCCAAAAACAAGUCUCAUUUUUCUUCGUGUGUAAAACUUGUGCUCUGCACCGAGCGCAAACCGGAGAGCACUGAAAA------- -------------------...(((((....(((..(((.........................((.....))((((((((......))))))))...)))..))))))))..------- ( -22.10, z-score = -1.21, R) >droEre2.scaffold_4690 10192902 110 - 18748788 GCCUUCAGAUGCAUAUGUGUCAUGUAUGUGACACCAUCGA--AAUAAGUCUCACUUUUCUCUGUGUGUAAAACUUGUGCUCUGUAACGAGCGCAAUUCGGAGAGCACAAAAA-------- ((((((...((((((((((((((....)))))))....((--...(((.....)))...)).)))))))....((((((((......))))))))...)))).)).......-------- ( -32.50, z-score = -2.64, R) >droYak2.chrX 19337167 113 - 21770863 GCCUUCAGAUGC-----UAAACUAAAUAUGAGACCAUCGG--AAGAAGUCUUACUUUUCUCUGUGUGUAAAACUUGUGCUCUGCAACGAGCGCAAUUCGGAGAGCGGAGAGCACGGAAAA ...(((...(((-----(..........((((((..((..--..)).))))))((.(((((((.((.....))((((((((......))))))))..))))))).))..))))..))).. ( -33.10, z-score = -1.60, R) >droSec1.super_8 2293550 92 - 3762037 -------------------GCCUUCAGAUUCUCCCAUCCA--AACAAGUCUCAUUUUUCUCCGUGUGUAAAACUUGUGCUCUGCAGCGAGCGCAAACCGGAGAGCACGGAAAA------- -------------------.....................--.................(((((((.......((((((((......))))))))........)))))))...------- ( -22.36, z-score = -0.98, R) >droSim1.chrU 2941927 92 - 15797150 -------------------GCCUACAGAUUCUCCCAUCCA--AACAAGUCUCAUUUUUCUCCGUGUGUAAAACUUGUGCUCUGCAGCGAGCGCAAACCGGAGAGCACGGAAAA------- -------------------.....................--.................(((((((.......((((((((......))))))))........)))))))...------- ( -22.36, z-score = -1.08, R) >apiMel3.GroupUn 394286701 110 + 399230636 --UCUUUCAUGAUUUUUAUCUCAUUAUAAAAGACCGUUCUAACGUACGUCU-AUAUUUUUUAUUAUACAUAUCACGUAGUACCACUUAAACGUAAUCAAGAAAUUGUUAAAGA------- --(((((.(..((((((.............((((.((.(....).))))))-.....................((((((.....))...)))).....))))))..).)))))------- ( -13.50, z-score = -0.41, R) >consensus ___________________GCCUUCAGAUUAUACCAUCCA__AACAAGUCUCAUUUUUCUCCGUGUGUAAAACUUGUGCUCUGCAACGAGCGCAAACCGGAGAGCACAGAAAA_______ ...........................................................(((((((.......((((((((......))))))))........))))))).......... (-13.73 = -14.04 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:57 2011