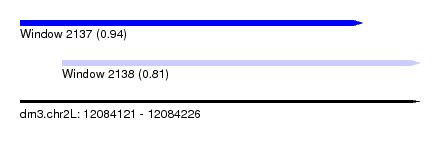
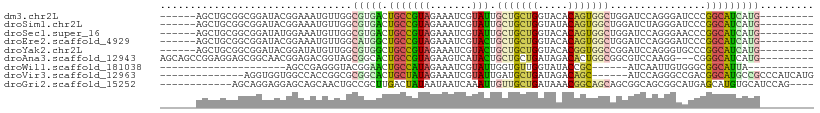
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,084,121 – 12,084,226 |
| Length | 105 |
| Max. P | 0.937290 |

| Location | 12,084,121 – 12,084,211 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 61.24 |
| Shannon entropy | 0.74825 |
| G+C content | 0.56213 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -10.20 |
| Energy contribution | -9.13 |
| Covariance contribution | -1.07 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
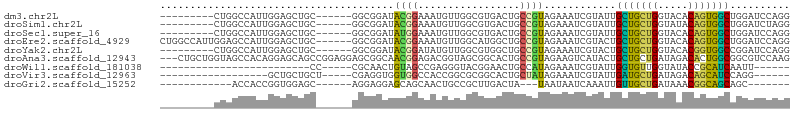
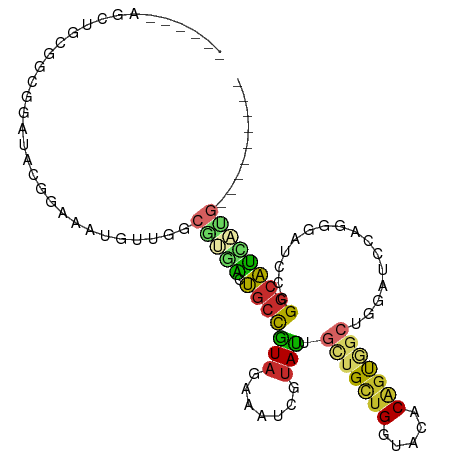
>dm3.chr2L 12084121 90 + 23011544 ---------CUGGCCAUUGGAGCUGC------GGCGGAUACGGAAAUGUUGGCGUGACUGCCGUAGAAAUCGUAUUGCUGCUGGUACACAGUGGCUGGAUCCAGG ---------(..(((((((((.((((------(((((.((((..........)))).)))))))))...))(((((......))))).)))))))..)....... ( -38.60, z-score = -3.35, R) >droSim1.chr2L 11881646 90 + 22036055 ---------CUGGCCAUUGGAGCUGC------GGCGGAUACGGAAAUGUUGGCGUGACUGCCGUAGAAAUCGUAUUGCUGCUGGUAUACAGUGGCUGGAUCUAGG ---------(..(((((((..(((((------((((.((((((......(((((....)))))......)))))))))))).)))...)))))))..)....... ( -37.00, z-score = -3.56, R) >droSec1.super_16 272349 90 + 1878335 ---------CUGGCCAUUGGAGCUGC------GGCGGAUAUGGAAAUGUUGGCGUGACUGCCGUAGAAAUCGUAUUGCUGCUGGUACACAGUGGCUGGAUCCAGG ---------(..(((((((((.((((------(((((.((((..........)))).)))))))))...))(((((......))))).)))))))..)....... ( -36.80, z-score = -2.91, R) >droEre2.scaffold_4929 13305309 99 - 26641161 CUGGCCAUUGGAGCCAUUGGAGCUGC------GGCGGAUACGGAAAUGUUGGCAUGGCUGCCGUAGAAAUCGUACUGCUGCUGGUACACAGUGGCUGGAUCCAGG (((((((.....((((((((.(((((------(((((.(((((......(((((....)))))......)))))))))))).))).).))))))))))..)))). ( -41.50, z-score = -2.44, R) >droYak2.chr2L 8500253 90 + 22324452 ---------CUGGCCAUUGGAGCUGC------GGCGGAUACGGAUAUGUUGGCGUGGCUGCCGUAGAAAUCGUACUGCUGCUGGUACACGGUGGCCGGAUCCAGG ---------((((((((((((.((((------(((((.((((..........)))).)))))))))...))(((((......))))).))))))))))....... ( -42.80, z-score = -3.31, R) >droAna3.scaffold_12943 2488694 102 - 5039921 ---CUGCUGGUAGCCACAGGAGCAGCCGGAGGAGCGGCAACGGAGACGGUAGCGGCACUGCCGUAGAAGUCAUACUGCUGCUGAUAGACACUGGCGGCGUCCAAG ---((((..((..(..(((.((((((((......))))......(((..(.(((((...))))).)..)))....)))).)))...)..))..))))........ ( -37.20, z-score = -0.03, R) >droWil1.scaffold_181038 28738 69 + 637489 -------------------------CC-----CGCAACUGUAGCCGAGGGUACGGAACUGCCAUAGAAAUCGUAUUGGUGUUGGUAUACCGCAUCAAUU------ -------------------------..-----.(((.(((((.(....).)))))...)))............((((((((.((....)))))))))).------ ( -17.10, z-score = -0.09, R) >droVir3.scaffold_12963 10204903 76 + 20206255 ------------------GCUGCUGCU-----CGAGGUGGUGGCCACCGGCGCGGCACUGCUAUAGAAAUCGUAUUGAUGCUGAUAGACAGCAUCCAGG------ ------------------((((((((.-----(..(((((...))))).).))))))..))...............(((((((.....)))))))....------ ( -27.60, z-score = -0.89, R) >droGri2.scaffold_15252 13335063 77 + 17193109 ------------ACCACCGGUGGAGC------AGGAGGAGCAGCAACUGCCGCUUGACUA---UAAUAAUCAAAUUGUUGCUGAUAAACGGCAGCAGC------- ------------......(((.((((------.(.((.........)).).)))).))).---...........(((((((((.....))))))))).------- ( -21.30, z-score = -0.73, R) >consensus _________CUGGCCAUUGGAGCUGC______GGCGGAUACGGAAAUGUUGGCGUGACUGCCGUAGAAAUCGUAUUGCUGCUGGUACACAGUGGCUGGAUCCAGG .......................................((((.................))))............(((((((.....))))))).......... (-10.20 = -9.13 + -1.07)
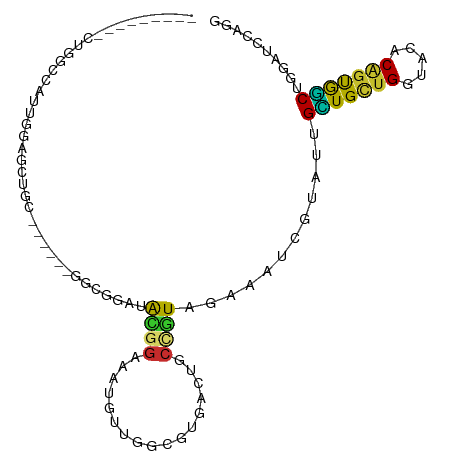
| Location | 12,084,132 – 12,084,226 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 60.97 |
| Shannon entropy | 0.76974 |
| G+C content | 0.56495 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -12.35 |
| Energy contribution | -12.79 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.88 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.811861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12084132 94 + 23011544 ------AGCUGCGGCGGAUACGGAAAUGUUGGCGUGACUGCCGUAGAAAUCGUAUUGCUGCUGGUACACAGUGGCUGGAUCCAGGGAUCCCGGCAUCAUG--------- ------..(((((((((.((((..........)))).)))))))))...(((....((..(((.....)))..)).(((((....)))))))).......--------- ( -36.70, z-score = -1.77, R) >droSim1.chr2L 11881657 94 + 22036055 ------AGCUGCGGCGGAUACGGAAAUGUUGGCGUGACUGCCGUAGAAAUCGUAUUGCUGCUGGUAUACAGUGGCUGGAUCUAGGGAUCCCGGCAUCAUG--------- ------..(((((((((.((((..........)))).)))))))))...(((....((..(((.....)))..)).(((((....)))))))).......--------- ( -35.60, z-score = -1.97, R) >droSec1.super_16 272360 94 + 1878335 ------AGCUGCGGCGGAUAUGGAAAUGUUGGCGUGACUGCCGUAGAAAUCGUAUUGCUGCUGGUACACAGUGGCUGGAUCCAGGGAACCCGGCAUCAUG--------- ------..(((((((((.((((..........)))).)))))))))..........((..(((.....)))..))..(((((.((....)))).)))...--------- ( -33.40, z-score = -1.03, R) >droEre2.scaffold_4929 13305329 94 - 26641161 ------AGCUGCGGCGGAUACGGAAAUGUUGGCAUGGCUGCCGUAGAAAUCGUACUGCUGCUGGUACACAGUGGCUGGAUCCAGGGAUCCCGGCAUCAUG--------- ------.((((((((((.(((((......(((((....)))))......)))))))))))).))).((..(((.(.(((((....))))).).)))..))--------- ( -36.00, z-score = -1.17, R) >droYak2.chr2L 8500264 94 + 22324452 ------AGCUGCGGCGGAUACGGAUAUGUUGGCGUGGCUGCCGUAGAAAUCGUACUGCUGCUGGUACACGGUGGCCGGAUCCAGGGUGCCCGGCAUCAUG--------- ------.((((.((((....)....((.((((..((((..((((.(....)(((((......)))))))))..))))...)))).)))))))))......--------- ( -35.90, z-score = 0.19, R) >droAna3.scaffold_12943 2488711 97 - 5039921 AGCAGCCGGAGGAGCGGCAACGGAGACGGUAGCGGCACUGCCGUAGAAGUCAUACUGCUGCUGAUAGACACUGGCGGCGUCCAAGG---CGGGCAUCAUG--------- .((.(((((.(...(((((.(((.(((..(.(((((...))))).)..)))...))).))))).....).))))).))((((....---.))))......--------- ( -36.50, z-score = -0.63, R) >droWil1.scaffold_181038 28749 71 + 637489 ---------------------AGCCGAGGGUACGGAACUGCCAUAGAAAUCGUAUUGGUGUUGGUAUACCGC------AUCAAUUGUGGGCGGCAUUA----------- ---------------------.((((...((((.((...((((..(....)....)))).)).)))).((((------(.....))))).))))....----------- ( -21.60, z-score = -0.95, R) >droVir3.scaffold_12963 10204914 89 + 20206255 --------------AGGUGGUGGCCACCGGCGCGGCACUGCUAUAGAAAUCGUAUUGAUGCUGAUAGACAGC------AUCCAGGGCCGACGGCAUGCCGCCCAUCAUG --------------.(((((((((..(((...((((.(((.....(....).....(((((((.....))))------)))))).)))).)))...)))).)))))... ( -36.80, z-score = -1.71, R) >droGri2.scaffold_15252 13335074 93 + 17193109 ------------AGCAGGAGGAGCAGCAACUGCCGCUUGACUAUAAUAAUCAAAUUGUUGCUGAUAAACGGCAGCAGCGGCAGCGGCAUGAGCAUGUGCAUCCAG---- ------------.(((.(..(..((((..(((((((((((.........))))..((((((((.....)))))))))))))))..)).))..).).)))......---- ( -34.40, z-score = -1.98, R) >consensus ______AGCUGCGGCGGAUACGGAAAUGUUGGCGUGACUGCCGUAGAAAUCGUAUUGCUGCUGGUACACAGUGGCUGGAUCCAGGGAUCCCGGCAUCAUG_________ .............((.((((......)))).))((((.(((((((.......))).(((((((.....)))))))................)))))))).......... (-12.35 = -12.79 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:23 2011