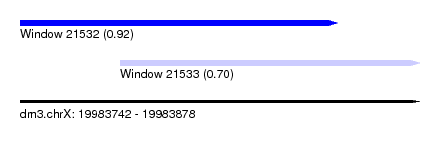
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,983,742 – 19,983,878 |
| Length | 136 |
| Max. P | 0.915672 |

| Location | 19,983,742 – 19,983,850 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.13 |
| Shannon entropy | 0.14854 |
| G+C content | 0.53056 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -34.50 |
| Energy contribution | -34.74 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.915672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
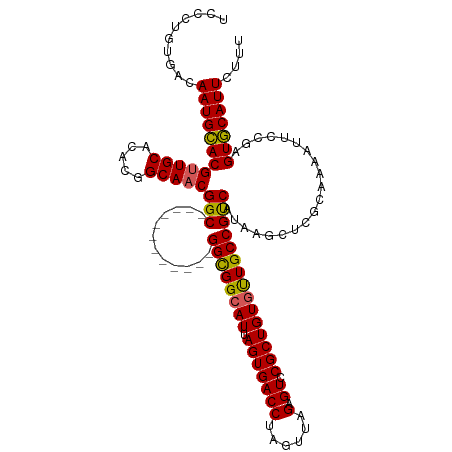
>dm3.chrX 19983742 108 + 22422827 UCCCUGUGACAAUGUACGUUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGUUGCCGUCAUAAGCUCGCAAAAUUCCGAGUACAUUCUUU ....((((((...((.((((((.....))))))))------------((((((((.((((((((....)).)).)))))))))))))))))).(((((........)))))......... ( -37.70, z-score = -2.49, R) >droSim1.chrX 15369479 108 + 17042790 UCCCUGUGACAAUGCACGUUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGCUGCCGUCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUU ....((((((...((.((((((.....))))))))------------((((((((.((((((((....)).)).)))))))))))))))))).(((((........)))))......... ( -42.90, z-score = -3.11, R) >droSec1.super_8 2209510 108 + 3762037 UCCCUGUGACAAUGCACGUUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGCUGCCGUCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUU ....((((((...((.((((((.....))))))))------------((((((((.((((((((....)).)).)))))))))))))))))).(((((........)))))......... ( -42.90, z-score = -3.11, R) >droYak2.chrX 10339857 120 - 21770863 UCCCUGUGACAAUGCACGUUGCACGUGGCAACGGCAACGGUGGCGUCGGUGGCAUUAGUGACCUAGUUUGUGUCCGCUGUGUUGCCGCCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUU ..........((((((((((((.(((....)))))))).(((((...(((((((.((((((((......).)).)))))...)))))))....)).)))..........))))))).... ( -43.60, z-score = -1.10, R) >droEre2.scaffold_4690 10126747 108 + 18748788 UCCCUGUGACAAUGCACGCUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUUGAGUCCGCUGUAUUGCCGUCAUAAGCUGGCAAAAUUCCGAGUGCAUUCUUU ..........((((((((((....(((((((..((------------(((((.(((...(((...)))..)))))))))).)))))))....)))(((.......))).))))))).... ( -35.90, z-score = -0.94, R) >consensus UCCCUGUGACAAUGCACGUUGCACACGGCAACGGC____________GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGUUGCCGUCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUU ....((((((...((.((((((.....))))))))............((((((((.(((((((......).)).)))))))))))))))))).(((((........)))))......... (-34.50 = -34.74 + 0.24)

| Location | 19,983,776 – 19,983,878 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.82 |
| Shannon entropy | 0.19813 |
| G+C content | 0.49195 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -23.55 |
| Energy contribution | -23.43 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
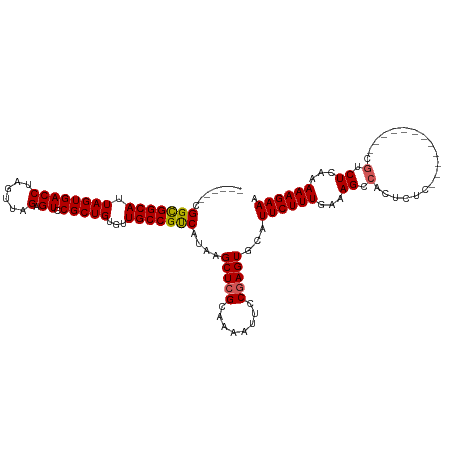
>dm3.chrX 19983776 102 + 22422827 ------CGGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGUUGCCGUCAUAAGCUCGCAAAAUUCCGAGUACAUUCUUUGAAAGCCACUCAC------------CGACUCAAAAAGAAA ------.(((((((.(((((((((....)).)).)))))...)))))))....(((((........)))))...((((((...((.(......------------.).))...)))))). ( -26.60, z-score = -1.62, R) >droSim1.chrX 15369513 102 + 17042790 ------CGGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGCUGCCGUCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUUGAAAGCCACUCUC------------CGUCUCAAAAAGAAA ------(((((((((.((((((((....)).)).)))))))))))))......(((((........)))))...((((((...((.(......------------.).))...)))))). ( -28.40, z-score = -1.60, R) >droSec1.super_8 2209544 102 + 3762037 ------CGGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGCUGCCGUCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUUGAAAGCCACUCUC------------CGUCUCAAAAAGAAA ------(((((((((.((((((((....)).)).)))))))))))))......(((((........)))))...((((((...((.(......------------.).))...)))))). ( -28.40, z-score = -1.60, R) >droYak2.chrX 10339897 108 - 21770863 UGGCGUCGGUGGCAUUAGUGACCUAGUUUGUGUCCGCUGUGUUGCCGCCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUUGAAAGCCACUCUC------------CGUCUGAAAAAGAAC ((((...(((((((.((((((((......).)).)))))...)))))))....(((((........))))).............)))).....------------..(((.....))).. ( -31.10, z-score = -1.48, R) >droEre2.scaffold_4690 10126781 114 + 18748788 ------CGGCGGCAUUAGUGACCUAGUUUGAGUCCGCUGUAUUGCCGUCAUAAGCUGGCAAAAUUCCGAGUGCAUUCUUUGAAAGCCACUCCCCAGCCCCAUCCCCAUCUCAAAAAGAAC ------.(((((((.(((((..((......))..)))))...)))))))....(((((.........((((((.(((...))).).))))).)))))....................... ( -30.10, z-score = -2.20, R) >consensus ______CGGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGUUGCCGUCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUUGAAAGCCACUCUC____________CGUCUCAAAAAGAAA .......(((((((.((((((((......).)).)))))...)))))))....(((((........)))))...((((((.................................)))))). (-23.55 = -23.43 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:49 2011