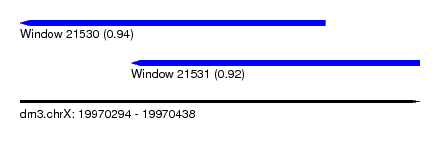
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,970,294 – 19,970,438 |
| Length | 144 |
| Max. P | 0.935636 |

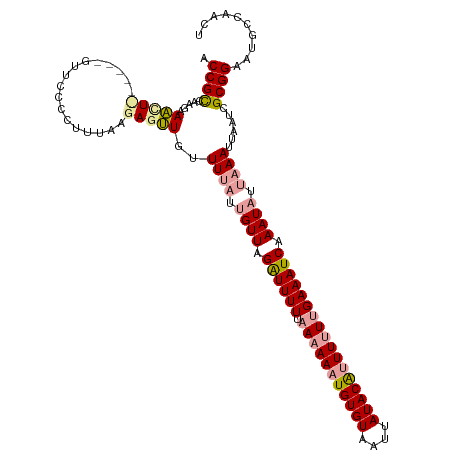
| Location | 19,970,294 – 19,970,404 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Shannon entropy | 0.24125 |
| G+C content | 0.30159 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -17.48 |
| Energy contribution | -18.93 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19970294 110 - 22422827 --UUUGUUAGGUUUUU-AAAAAUGUGUAAUCAUACGGUUUUUGAAAUCAAAU---UAAUCG--CGCGGAAUGUCAAUUUGUUGUACUCCGUGAAAGCCUUGCAUUAACGACACGACAU --..(((.(((((((.-.....((((....))))(((((((((....)))).---.)))))--((((((.((.(((....))))).))))))))))))).)))............... ( -22.20, z-score = -0.21, R) >droSec1.super_8 2196036 118 - 3762037 UUAUUGUUAGAUUUUUAAAAAAUGUGUAAUUAUACAUUUUUUGAAAUCAAAUAUUAAAUUAAUCGCGGAGUGCCAACUCUGUGUACAUCGUUAAAGUCUUGCAUUAACAACGCGACAU (((.((((.((((((.(((((((((((....))))))))))))))))).)))).))).....(((((((((((((....)).)))).))(((((.(.....).)))))..)))))... ( -30.00, z-score = -3.94, R) >droSim1.chrX_random 5179146 118 - 5698898 UUAUUGUUAGAUUUUUAAAAAAUGUGUAAUUAUACAUUUCUUGAAAUCAAAUAUUAAAUUAAUCGCGGAAUGCCAACACUUUGUACUUCCUUAAAGCCUUGCAUUAACAACGCGACAU (((.((((.((((((....((((((((....))))))))...)))))).)))).))).....(((((.(((((.((..(((((........)))))..))))))).....)))))... ( -25.10, z-score = -3.58, R) >consensus UUAUUGUUAGAUUUUUAAAAAAUGUGUAAUUAUACAUUUUUUGAAAUCAAAUAUUAAAUUAAUCGCGGAAUGCCAACUCUUUGUACUUCGUUAAAGCCUUGCAUUAACAACGCGACAU ....((((.((((((.(((((((((((....)))))))))))))))))................(((.((((.(((..(((((........)))))..))))))).....))))))). (-17.48 = -18.93 + 1.45)

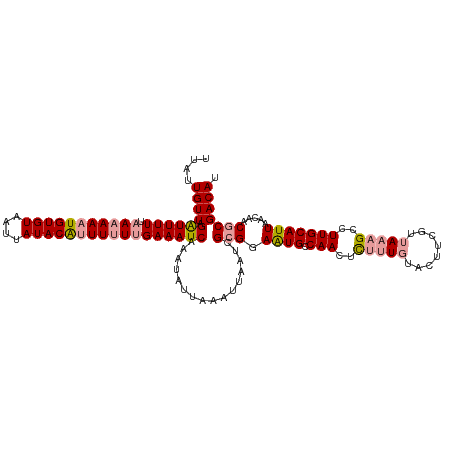
| Location | 19,970,334 – 19,970,438 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Shannon entropy | 0.29359 |
| G+C content | 0.31185 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -15.70 |
| Energy contribution | -16.80 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19970334 104 - 22422827 ACCGUCAACCACUU------GUUCCCCAUUAAGAGGUGUUUU--UGUUAGGUUUUU-AAAAAUGUGUAAUCAUACGGUUUUUGAAAUCAAAUU---AAUCG--CGCGGAAUGUCAAUU .((((..(((.(((------(........)))).)))((..(--((....(.((((-((((((((((....)))).)))))))))).)....)---))..)--))))).......... ( -18.40, z-score = -0.07, R) >droSec1.super_8 2196076 113 - 3762037 ACCGCCAAGAAACUC-----GUUCCCCUUCCAGAGUUGUUUUAUUGUUAGAUUUUUAAAAAAUGUGUAAUUAUACAUUUUUUGAAAUCAAAUAUUAAAUUAAUCGCGGAGUGCCAACU .((((.....(((((-----............)))))..((((.((((.((((((.(((((((((((....))))))))))))))))).)))).))))......)))).......... ( -26.20, z-score = -4.16, R) >droSim1.chrX_random 5179186 118 - 5698898 ACCGCCAAGAAACUUUAAGAGUUGCCCUUUAAGACUUGUUUUAUUGUUAGAUUUUUAAAAAAUGUGUAAUUAUACAUUUCUUGAAAUCAAAUAUUAAAUUAAUCGCGGAAUGCCAACA .((((((((...(((.(((.......))).))).)))).((((.((((.((((((....((((((((....))))))))...)))))).)))).))))......)))).......... ( -22.40, z-score = -2.36, R) >consensus ACCGCCAAGAAACU______GUUCCCCUUUAAGAGUUGUUUUAUUGUUAGAUUUUUAAAAAAUGUGUAAUUAUACAUUUUUUGAAAUCAAAUAUUAAAUUAAUCGCGGAAUGCCAACU .((((.....(((((.................)))))..((((.((((.((((((.(((((((((((....))))))))))))))))).)))).))))......)))).......... (-15.70 = -16.80 + 1.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:47 2011