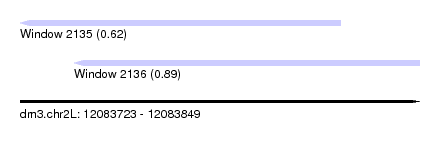
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,083,723 – 12,083,849 |
| Length | 126 |
| Max. P | 0.886046 |

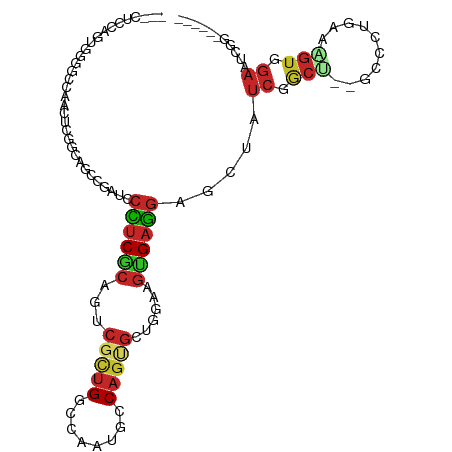
| Location | 12,083,723 – 12,083,824 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 65.41 |
| Shannon entropy | 0.70448 |
| G+C content | 0.62021 |
| Mean single sequence MFE | -39.13 |
| Consensus MFE | -11.83 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
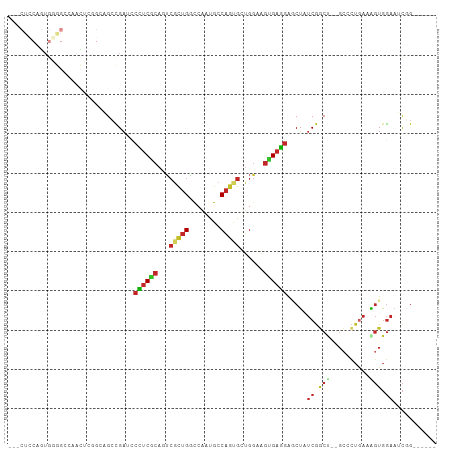
>dm3.chr2L 12083723 101 - 23011544 CUCCUCCAGUGGGGCCAACUCGGGAGCCGAUCCCUCGCAGUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAGCUAUCGGCU--GCCCUGAAAGUGGAAUCGG------ ((((......))))(((..(((((((((((((((((((((.(((((((....)))))))))....)))))).)..)))))))--.))).))...)))......------ ( -44.40, z-score = -1.59, R) >droSim1.chr2L 11881248 101 - 22036055 CUCCUCCAGUGGGGCCAACUCGGCAGCCGAUCCCUCGCAUUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAUCUAUCGGCU--GCCCUGAAAGUGGAAUCGG------ ....((((.((((.....)))((((((((((.((((((.(((((((((....))))))...))).))))))....)))))))--))).....).)))).....------ ( -45.50, z-score = -2.64, R) >droSec1.super_16 271951 101 - 1878335 CUCCUCCAGUGGGGCCAACUCGGCAGCCGAUCCCUCGCAUUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAGCUAUCGGCU--GCCCUGAAAGUGGAAUCGG------ ....((((.((((.....)))(((((((((((((((((.(((((((((....))))))...))).)))))).)..)))))))--))).....).)))).....------ ( -45.70, z-score = -2.31, R) >droEre2.scaffold_4929 13304914 98 + 26641161 ---CUCCAGUGGGGUCAACUCGGGAGCCGAUCCCUCGCAGUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAGCUAUCGUCU--GCCCUGAAGGUGGAAUCGG------ ---((((...(((.....))).))))(((((.((((((((.(((((((....)))))))))....))))))........((.--.((.....))..)))))))------ ( -38.40, z-score = -0.60, R) >droYak2.chr2L 8499858 98 - 22324452 ---CUCCAGUGGGGGCCACUCGGGAGCCGAUGCCUCGCAGUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAGCUAUCGGCC--GCCCUGAAGGUGGAAUCGG------ ---((((....))))(((((((((.(((((((((((((((.(((((((....)))))))))....))))))...))))))).--..)))))..))))......------ ( -44.40, z-score = -0.95, R) >droAna3.scaffold_12943 2488305 95 + 5039921 CUCCUCCGGUGCGGG------AGCCGCCGAUCCCUCACAGUCGCUGGCCAACGCCAGUGCUGGCAGUGAGGAGCUCUCGGCU--GCCCUUAAAGUGGAGACCG------ ...(((((.(..(((------.((.(((((.(((((((.(((((((((....))))))...))).)))))).)...))))).--)))))...).)))))....------ ( -43.90, z-score = -1.82, R) >dp4.chr4_group2 361063 77 + 1235136 ------------------------CGGCGAGUCCUCGCAGUCGUUGAUCAAUGCCAGCGGUGGCAGUGAGGAACUGUCGGCC--GCACUGAAAGUGGAGGCCG------ ------------------------.((((..(((((((.(((...)))...(((((....))))))))))))..))))((((--.(((.....)))..)))).------ ( -33.10, z-score = -1.93, R) >droPer1.super_8 3021992 77 - 3966273 ------------------------CGGCGAGUCCUCGCAGUCGUUGAUCAAUGCCAGCGGUGGCAGUGAGGAACUGUCGGCC--GCACUGAAAGUGGAGGCCG------ ------------------------.((((..(((((((.(((...)))...(((((....))))))))))))..))))((((--.(((.....)))..)))).------ ( -33.10, z-score = -1.93, R) >droWil1.scaffold_181038 28346 91 - 637489 -------AAUGCAACAAAUC---CAGCUGAGUCUUCGCAAUCACUGGCCAAUGCCAGCGUUGACAGCGAAGAUCUCUCUGCU--GAUUUAAAAGUGGAAGCAG------ -------..(((........---.....((((((((((..((((((((....)))))...)))..))))))).)))((..((--........))..)).))).------ ( -31.30, z-score = -2.87, R) >anoGam1.chrX 15834788 107 - 22145176 CGGCUACAGUGUAGUGGAUGC-GGAGCUGCCCCGCCUCACGAUUGGCUCGCUGUCGUCGCCGG-GCCGACUGUCGCCCAACCUGUUCCUGAACCUCAACUUUGACCAUC .(((.(((((...((((..((-((.......)))).))))...(((((((.((....)).)))-))))))))).)))................................ ( -31.50, z-score = 0.80, R) >consensus ___CUCCAGUGGGGCCAACUCGGCAGCCGAUCCCUCGCAGUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAGCUAUCGGCU__GCCCUGAAAGUGGAAUCGG______ ................................((((((...(((((........)))))......)))))).(((.((((.......)))).))).............. (-11.83 = -12.40 + 0.57)

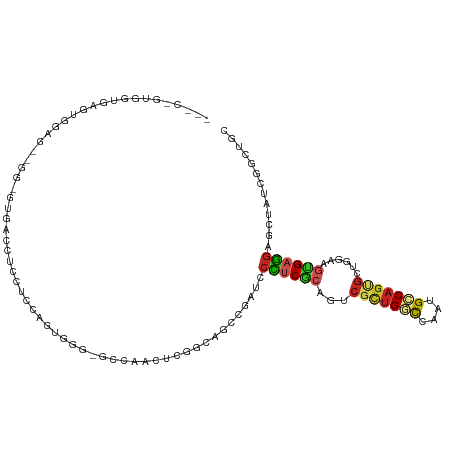
| Location | 12,083,740 – 12,083,849 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 57.95 |
| Shannon entropy | 0.90665 |
| G+C content | 0.65668 |
| Mean single sequence MFE | -44.64 |
| Consensus MFE | -13.79 |
| Energy contribution | -13.95 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12083740 109 - 23011544 ---CGGUGGUGAGUGGAGCUGGUGUGACCUCCUCCAGUGGG-GCCAACUCGGGAGCCGAUCCCUCGCAGUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAGCUAUCGGCUGC ---(((((((.(.(((((..((.....))..))))).)...-))).)).))(.((((((((((((((((.(((((((....)))))))))....)))))).)..))))))).) ( -46.20, z-score = -0.37, R) >droSim1.chr2L 11881265 109 - 22036055 ---CGGUGGUGAGUGGAGCUGGUGUGACCUCCUCCAGUGGG-GCCAACUCGGCAGCCGAUCCCUCGCAUUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAUCUAUCGGCUGC ---(((((((.(.(((((..((.....))..))))).)...-))).)).))(((((((((.((((((.(((((((((....))))))...))).))))))....))))))))) ( -49.50, z-score = -1.65, R) >droSec1.super_16 271968 109 - 1878335 ---CGGUGGUGAGUGGAGCUGGUGUGACCUCCUCCAGUGGG-GCCAACUCGGCAGCCGAUCCCUCGCAUUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAGCUAUCGGCUGC ---(((((((.(.(((((..((.....))..))))).)...-))).)).))((((((((((((((((.(((((((((....))))))...))).)))))).)..))))))))) ( -49.70, z-score = -1.41, R) >droEre2.scaffold_4929 13304931 106 + 26641161 ---CUGUGGUGAGUGGAGCUGCGGUGAG---CUCCAGUGGG-GUCAACUCGGGAGCCGAUCCCUCGCAGUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAGCUAUCGUCUGC ---..............((.(((((.((---((((.(.(((-(((..((....))..)))))).).(((.(((((((....)))))))))).......)))))))))))..)) ( -43.00, z-score = -0.46, R) >droYak2.chr2L 8499875 106 - 22324452 ---CUGUGGUGAGCGGAGCUGGAGUGAC---CUCCAGUGGG-GGCCACUCGGGAGCCGAUGCCUCGCAGUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAGCUAUCGGCCGC ---..(((((.(((((..((.(((((.(---((((...)))-)).))))).))..))....((((((((.(((((((....)))))))))....)))))).)))....))))) ( -52.10, z-score = -1.88, R) >droAna3.scaffold_12943 2488322 97 + 5039921 ------CCGUCGGCGGA---GGCCUGACCUCCUCCGGUGCG-GG------AGCCGCCGAUCCCUCACAGUCGCUGGCCAACGCCAGUGCUGGCAGUGAGGAGCUCUCGGCUGC ------(((((((.(((---((.....))))).)))..)))-).------.((.(((((.(((((((.(((((((((....))))))...))).)))))).)...))))).)) ( -49.30, z-score = -1.26, R) >dp4.chr4_group2 361080 85 + 1235136 ----------------------------CGGUUGUGGGUGGAGCCGGUCCCACCGGCGAGUCCUCGCAGUCGUUGAUCAAUGCCAGCGGUGGCAGUGAGGAACUGUCGGCCGC ----------------------------((..((((((.((.(((((.....)))))...))))))))..)).............(((((((((((.....)))))).))))) ( -38.80, z-score = -1.65, R) >droPer1.super_8 3022009 85 - 3966273 ----------------------------CGGUUGUGGGUGGAGCCGGUCCCACCGGCGAGUCCUCGCAGUCGUUGAUCAAUGCCAGCGGUGGCAGUGAGGAACUGUCGGCCGC ----------------------------((..((((((.((.(((((.....)))))...))))))))..)).............(((((((((((.....)))))).))))) ( -38.80, z-score = -1.65, R) >droWil1.scaffold_181038 28363 97 - 637489 CCGUAGUAGUGGGUGGAG--GUAGUAA-----UGCAACAAA-UC--------CAGCUGAGUCUUCGCAAUCACUGGCCAAUGCCAGCGUUGACAGCGAAGAUCUCUCUGCUGA ...((((((.((.((((.--(((....-----)))......-))--------)).))((((((((((..((((((((....)))))...)))..))))))).))).)))))). ( -36.70, z-score = -2.74, R) >droVir3.scaffold_12963 10204523 85 - 20206255 ----------------------------CCGCCGUGGUGGGUGGUGGCUCCGCCUCCGAGACCACGACCUCGCUGACCAAUGUCAGCGGCGGCAGCGAGGAGCUUAGCGCCGC ----------------------------.(((((((((.((.((((....)))).))...)))))).((((((((.((..((....))..))))))))))......))).... ( -40.40, z-score = -1.02, R) >anoGam1.chrX 15834817 103 - 22145176 ---CCGGCCCGGGUGUCGUCAGCUCGUCCGGCUACAGUGU--AGUGGAUGCGGAGCUGCCCCGCCUCACGAUUGGCUCGCUGUCGUCGCCGG-GCCGACUGUCGCCCAA---- ---.((((((((.((.((.((((.((((((.((((...))--))))))))..((((((...((.....))..)))))))))).)).))))))-))))............---- ( -46.50, z-score = -0.70, R) >consensus ___C_GUGGUGAGUGGAG__GG_GUGACCUCCUCCAGUGGG_GCCAACUCGGCAGCCGAUCCCUCGCAGUCGCUGGCCAAUGCCAGUGCUGGAAGUGAGGAGCUAUCGGCUGC .............................................................((((((...(((((((....)))))))......))))))............. (-13.79 = -13.95 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:21 2011