
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,912,717 – 19,912,868 |
| Length | 151 |
| Max. P | 0.672636 |

| Location | 19,912,717 – 19,912,868 |
|---|---|
| Length | 151 |
| Sequences | 4 |
| Columns | 155 |
| Reading direction | reverse |
| Mean pairwise identity | 86.15 |
| Shannon entropy | 0.22075 |
| G+C content | 0.48150 |
| Mean single sequence MFE | -43.39 |
| Consensus MFE | -38.67 |
| Energy contribution | -38.05 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
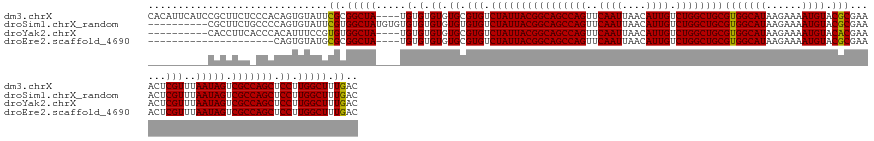
>dm3.chrX 19912717 151 - 22422827 CACAUUCAUCCGCUUCUCCCACAGUGUAUUCGCGGCUA----UGUGUGUGUGCGUGUCUAUUACGGCAGCCAGUUCAAUUAACAUUGUCUGGCUGCGUGGCAUAAGAAAAUGUACGCGAAACUCGUUUAAUAGUCGCCAGCUCCUUGGCUUUGAC (((((.(((((((..................))))..)----)).)))))((.(((.((((((((((((((((..((((....)))).))))))))(((((((......)))).)))......)))..))))).)))))(((....)))...... ( -43.57, z-score = -0.86, R) >droSim1.chrX_random 5165718 145 - 5698898 ----------CGCUUCUGCCCCAGUGUAUUCGUGGCUAUGUGUGUGUGUGUGUGUGUCUAUUACGGCAGCCAGUUCAAUUAACAUUGUCUGGCUGCGUGGCAUAAGAAAAUGUACGCGAAACUCGUUUAAUAGUCGCCAGCUCCUUGGCUUUGAC ----------....((.(((..((.(..((.(((((((((((((..(.....((((((....(((.(((((((..((((....)))).)))))))))))))))).....)..)))))(.....).....)))))))).))..))).)))...)). ( -42.30, z-score = -0.56, R) >droYak2.chrX 10273179 141 + 21770863 ----------CACCUUCACCCACAUUUCCGUGUGGCUA----UGUGUGUGUGCGUGUCUAUUACGGCAGCCAGUUCAAUUAACAUUGUCUGGCUGCGUGGCAUAAGAAAAUGUACACGAAACUCGUUUAAUAGUCGCCAGCUCCUUGGCUUUGAC ----------(((...(((.(((((..((....))..)----)))).)))...))).((((((((((((((((..((((....)))).))))))))(((((((......)))).)))......)))..)))))..(((((....)))))...... ( -44.20, z-score = -2.04, R) >droEre2.scaffold_4690 10062324 130 - 18748788 ---------------------CAGUGUAUGCGCGGCUA----UGUGUGUGUGCGUGUCUAUUACGGCAGCCAGUUCAAUUAACAUUGUCUGGCUGCGUGGCAUAAGAAAAUGUACGCGAAACUCGUUUAAUAGUCGCCAGCUCCUUGGCUUUGAC ---------------------..(..((((((((....----))))))))..)..(.((((((((((((((((..((((....)))).))))))))(((((((......)))).)))......)))..))))).)(((((....)))))...... ( -43.50, z-score = -1.36, R) >consensus __________CGCUUCU_CCCCAGUGUAUGCGCGGCUA____UGUGUGUGUGCGUGUCUAUUACGGCAGCCAGUUCAAUUAACAUUGUCUGGCUGCGUGGCAUAAGAAAAUGUACGCGAAACUCGUUUAAUAGUCGCCAGCUCCUUGGCUUUGAC ...............................(((((((....((.((((((((((.(((.(((((.(((((((..((((....)))).))))))))))))....)))..))))))))...)).)).....))))))).((((....))))..... (-38.67 = -38.05 + -0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:40 2011