
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,891,545 – 19,891,716 |
| Length | 171 |
| Max. P | 0.833653 |

| Location | 19,891,545 – 19,891,716 |
|---|---|
| Length | 171 |
| Sequences | 6 |
| Columns | 178 |
| Reading direction | reverse |
| Mean pairwise identity | 54.61 |
| Shannon entropy | 0.79352 |
| G+C content | 0.48041 |
| Mean single sequence MFE | -52.87 |
| Consensus MFE | -9.20 |
| Energy contribution | -12.93 |
| Covariance contribution | 3.73 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.17 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.833653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
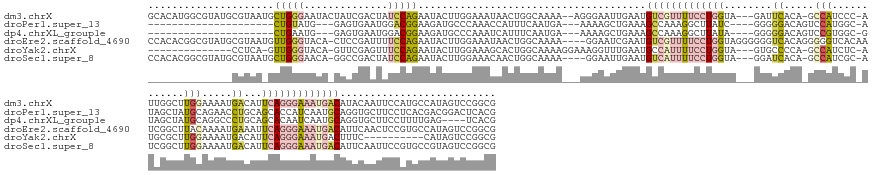
>dm3.chrX 19891545 171 - 22422827 GCACAUGGCGUAUGCGUAAUGCUGGGAAUACUAUCGACUAUCCAGAAUACUUGGAAAUAACUGGCAAAA--AGGGAAUUGAAUGUCGUUUUCCUGGUA---GAUUCACA-GCCAUCCC-AUUGGCUUGGAAAAUGACAUUCAGGGAAAUGACAUACAAUUCCAUGCCAUAGUCCGGCG .......(((....)))...((((.((......))((((((((((.....)))))......(((((...--..(((((((.(((((((((((((((((---..(((..(-((((....-..)))))..)))..))....))))))))))))))).))))))).)))))))))))))). ( -59.60, z-score = -4.18, R) >droPer1.super_13 1223252 146 - 2293547 ---------------------CUGUAUG---GAGUGAAUGGACGGAAGAUGCCCAAACCAUUUCAAUGA---AAAAGCUGAAAGCCAAAGGCUUAUC----GGGGGACAGUCCAUGGC-AUAGCUAUGCAGAACCUGCAGCACCAUCAAUGCAGGUGCUUCCUCACGACGGACUCACG ---------------------(((((((---(.(((.((((((........(((....(((....))).---.....(((((((((...))))).))----)))))...))))))..)-))..)))))))).(((((((..........)))))))...(((.......)))...... ( -44.30, z-score = -0.99, R) >dp4.chrXL_group1e 5196881 142 + 12523060 ---------------------CUGAAUG---GAGUGAAUGGACGGAAGAUGCCCAAAUCAUUUCAAUGA---AAAAGCUGAAAGCCAAAGGCUUAUA----GGGGGACAGUCCGUGGC-GUAGCUAUGCAGGCCCUGCAGCACAAUCAAUGCAGGUGCUUCCUUUUGAG----UCACG ---------------------......(---((.((......(....)...(((...((((....))))---.....(((.(((((...))))).))----))))..)).)))(((((-(((....)))((((((((((..........)))))).))))........)----)))). ( -42.80, z-score = -0.32, R) >droEre2.scaffold_4690 10043026 173 - 18748788 CCACACGGCGUAUGCGUAAUGUUGGGUACA-CUCCGAUUUUCCAGAAUACUUGGAAAUAACUGGCAAAA----GGAAUCGAAUGUCGUUUUCCUGGUAGGGGGGUCACAGGGGGUCACAAUCGGCUUACAAAAUGAAAUUCAGGGAAAUGACAUUCAACUCCGUGCCAUAGUCCGGCG (((((..(((....)))..)).))).....-..(((..(((((((.....)))))))....(((((...----(((...(((((((((((((((((....(......).(..((((......))))..)..........)))))))))))))))))...))).))))).....))).. ( -55.40, z-score = -2.02, R) >droYak2.chrX 10238440 147 + 21770863 --------------CCUCA-GUUGGGUACA-GUUCGAGUUUCCAGAAUACUUGGAAAGCACUGGCAAAAGGAAAGGUUUGAAUGCCAUUUUCCUGGUA---GUGCCCCA-GCCAUCUC-AUGCGCUUGGAAAAUGACAUUCAGGGAAAUGACUUUC----------CAUAGUCCGGCG --------------.....-((((((..((-((.(...(((((((.....)))))))).))))......((((((.((((((((.((((((((....(---((((...(-(....)).-..))))).)))))))).)))))))).......)))))----------)....)))))). ( -43.90, z-score = -0.77, R) >droSec1.super_8 2122413 168 - 3762037 CCACACGGCGUAUGCGUAAUGCUGGGAACA-GGCCGACUAUCCAGAAUACUUGGAAACAACUGGCAAAA----GGAAUUGAAUGUCAUUUUCCUGGUA---GGAUCACA-GCCAUCGC-AUCGGCUUGGAAAAUGACAUUCAGGGAAAUGACAUUCAAUUCCGUGCCGUAGUCCGGCG (((((..(((....)))..)).))).....-.(((((((((.........(((....)))..((((...----(((((((((((((((((((((((..---.(.(((..-.(((..((-....)).)))....))))..))))))))))))))))))))))).))))))))).)))). ( -71.20, z-score = -6.25, R) >consensus ______________CGUAA_GCUGGGUACA_GACCGAAUAUCCAGAAUACUUGGAAAUAACUGGCAAAA___AAGAAUUGAAUGCCAUUUUCCUGGUA___GGGUCACA_GCCAUCGC_AUAGGCUUGCAAAAUGACAUUCAGGGAAAUGACAUUCAAUUCCGUGCCAUAGUCCGGCG ....................((((((..............(((((.....)))))............................(((((((((((((...........................................)))))))))))))...................)))))). ( -9.20 = -12.93 + 3.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:36 2011