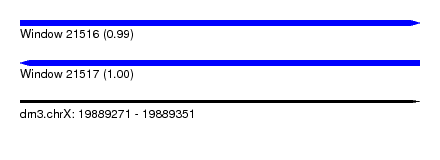
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,889,271 – 19,889,351 |
| Length | 80 |
| Max. P | 0.996881 |

| Location | 19,889,271 – 19,889,351 |
|---|---|
| Length | 80 |
| Sequences | 10 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 83.57 |
| Shannon entropy | 0.37517 |
| G+C content | 0.39277 |
| Mean single sequence MFE | -20.12 |
| Consensus MFE | -15.72 |
| Energy contribution | -15.95 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.992952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
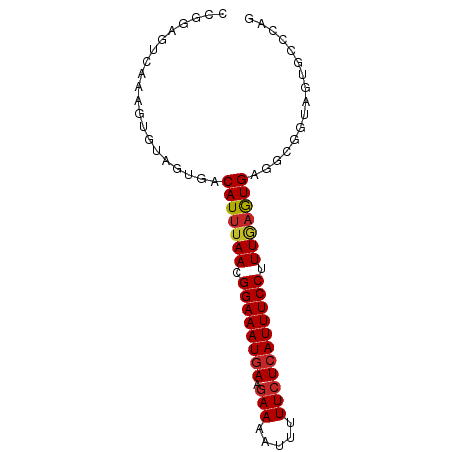
>dm3.chrX 19889271 80 + 22422827 CCGGAGUCAAAGUGUAGUGACAUUUAACGGAAAUGAAGAAAAUUUUUCUCAUUUCCUUUGAGUGAGGCGGUAGUGCCCAG .....((((........))))((((((.((((((((.(((.....))))))))))).))))))(.((((....))))).. ( -21.30, z-score = -2.07, R) >droWil1.scaffold_180777 3941482 80 - 4753960 CCAAAGUCAAAGUGUAGUGACAUUUAACGGAAAUGAAGAAAAUUUUUCUCAUUUCCUUUGAAUGUUGGAUGAAUGGACAA .....(((....(((..((((((((((.((((((((.(((.....))))))))))).)))))))))).)))....))).. ( -23.30, z-score = -3.44, R) >droPer1.super_13 1220295 68 + 2293547 CCAGAGUCAAAGUGUAGUGACAUUUAACGGAAAUGAAGAAAAUUUUUCUCAUUUCCUUUGAGUGAGCC------------ .....((((........))))((((((.((((((((.(((.....))))))))))).)))))).....------------ ( -17.30, z-score = -2.70, R) >dp4.chrXL_group1e 5193946 68 - 12523060 CCAGAGUCAAAGUGUAGUGACAUUUAACGGAAAUGAAGAAAAUUUUUCUCAUUUCCUUUGAGUGAGCC------------ .....((((........))))((((((.((((((((.(((.....))))))))))).)))))).....------------ ( -17.30, z-score = -2.70, R) >droAna3.scaffold_13417 5053369 77 - 6960332 CCAGAGUCAAAGUGUAGUGACAUUUAACGGAAAUGAAGAAAAUUUUUCUCAUUUCCUUUGAGUGA-GGCCG--UGCCCAG ...........(((..((..(((((((.((((((((.(((.....))))))))))).))))))).-.))..--))).... ( -20.30, z-score = -2.67, R) >droEre2.scaffold_4690 10040838 80 + 18748788 UCGGAGUCAAAGUGUAGUGACAUUUAACGGAAAUGAAGAAAAUUUUUCUCAUUUCCUUUGAGUGAGGCGGUAGUGCCCAA .....((((........))))((((((.((((((((.(((.....))))))))))).))))))(.((((....))))).. ( -21.30, z-score = -2.00, R) >droYak2.chrX 10236127 80 - 21770863 CUGGAGUCAAAGUGUAGUGACAUUUAACGGAAAUGAAGAAAAUUUUUCUCAUUUCCUUUGAGUGAGGCGGUAGUGCCCAG ((((.(.((.......((..(((((((.((((((((.(((.....))))))))))).)))))))..)).....))))))) ( -23.10, z-score = -2.65, R) >droSec1.super_8 2120213 80 + 3762037 CCGGAGUCAAAGUGUAGUGACAUUUAACGGAAAUGAAGAAAAUUUUUCUCAUUUCCUUUAAGUGAGGCGGUAGUGCCCAG .....((((........))))((((((.((((((((.(((.....))))))))))).))))))(.((((....))))).. ( -21.20, z-score = -2.37, R) >droSim1.chrX 15305192 80 + 17042790 CCGGAGUCAAAGUGUAGUGACAUUUAACGGAAAUGAAGAAAAUUUUUCUCAUUUCCUUUGAGUGAGGCGGUAGUGCCCAG .....((((........))))((((((.((((((((.(((.....))))))))))).))))))(.((((....))))).. ( -21.30, z-score = -2.07, R) >droGri2.scaffold_14853 2842917 69 + 10151454 ----------AGUGUAGCGACAUUUAAAGGAAAUUAACAAAUAUUUUCUCAUUUCCUUUC-GUGAGGCGUUCGCAUGCAG ----------..(((((((((....(((((((((................)))))))))(-....)..).)))).)))). ( -14.79, z-score = -1.67, R) >consensus CCGGAGUCAAAGUGUAGUGACAUUUAACGGAAAUGAAGAAAAUUUUUCUCAUUUCCUUUGAGUGAGGCGGUAGUGCCCAG ....................(((((((.((((((((.(((.....))))))))))).)))))))................ (-15.72 = -15.95 + 0.23)

| Location | 19,889,271 – 19,889,351 |
|---|---|
| Length | 80 |
| Sequences | 10 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 83.57 |
| Shannon entropy | 0.37517 |
| G+C content | 0.39277 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -12.62 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.996881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
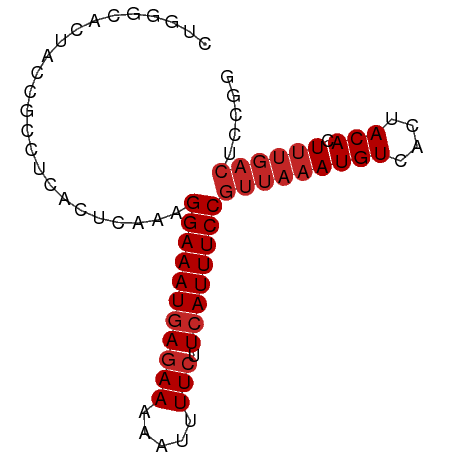
>dm3.chrX 19889271 80 - 22422827 CUGGGCACUACCGCCUCACUCAAAGGAAAUGAGAAAAAUUUUCUUCAUUUCCGUUAAAUGUCACUACACUUUGACUCCGG ..((((......))))........(((((((((((.....))).))))))))(((((((((....))).))))))..... ( -19.80, z-score = -3.36, R) >droWil1.scaffold_180777 3941482 80 + 4753960 UUGUCCAUUCAUCCAACAUUCAAAGGAAAUGAGAAAAAUUUUCUUCAUUUCCGUUAAAUGUCACUACACUUUGACUUUGG ............((((........(((((((((((.....))).))))))))(((((((((....))).)))))).)))) ( -17.10, z-score = -2.66, R) >droPer1.super_13 1220295 68 - 2293547 ------------GGCUCACUCAAAGGAAAUGAGAAAAAUUUUCUUCAUUUCCGUUAAAUGUCACUACACUUUGACUCUGG ------------............(((((((((((.....))).))))))))(((((((((....))).))))))..... ( -15.10, z-score = -2.69, R) >dp4.chrXL_group1e 5193946 68 + 12523060 ------------GGCUCACUCAAAGGAAAUGAGAAAAAUUUUCUUCAUUUCCGUUAAAUGUCACUACACUUUGACUCUGG ------------............(((((((((((.....))).))))))))(((((((((....))).))))))..... ( -15.10, z-score = -2.69, R) >droAna3.scaffold_13417 5053369 77 + 6960332 CUGGGCA--CGGCC-UCACUCAAAGGAAAUGAGAAAAAUUUUCUUCAUUUCCGUUAAAUGUCACUACACUUUGACUCUGG ..((((.--..)))-)........(((((((((((.....))).))))))))(((((((((....))).))))))..... ( -18.00, z-score = -2.33, R) >droEre2.scaffold_4690 10040838 80 - 18748788 UUGGGCACUACCGCCUCACUCAAAGGAAAUGAGAAAAAUUUUCUUCAUUUCCGUUAAAUGUCACUACACUUUGACUCCGA ..((((......))))........(((((((((((.....))).))))))))(((((((((....))).))))))..... ( -19.80, z-score = -3.35, R) >droYak2.chrX 10236127 80 + 21770863 CUGGGCACUACCGCCUCACUCAAAGGAAAUGAGAAAAAUUUUCUUCAUUUCCGUUAAAUGUCACUACACUUUGACUCCAG ((((((......))).........(((((((((((.....))).))))))))(((((((((....))).))))))..))) ( -19.90, z-score = -3.71, R) >droSec1.super_8 2120213 80 - 3762037 CUGGGCACUACCGCCUCACUUAAAGGAAAUGAGAAAAAUUUUCUUCAUUUCCGUUAAAUGUCACUACACUUUGACUCCGG ..((((......))))........(((((((((((.....))).))))))))(((((((((....))).))))))..... ( -19.80, z-score = -3.45, R) >droSim1.chrX 15305192 80 - 17042790 CUGGGCACUACCGCCUCACUCAAAGGAAAUGAGAAAAAUUUUCUUCAUUUCCGUUAAAUGUCACUACACUUUGACUCCGG ..((((......))))........(((((((((((.....))).))))))))(((((((((....))).))))))..... ( -19.80, z-score = -3.36, R) >droGri2.scaffold_14853 2842917 69 - 10151454 CUGCAUGCGAACGCCUCAC-GAAAGGAAAUGAGAAAAUAUUUGUUAAUUUCCUUUAAAUGUCGCUACACU---------- ......((((.((.....)-)(((((((((..(((....)))....))))))))).....))))......---------- ( -12.60, z-score = -1.47, R) >consensus CUGGGCACUACCGCCUCACUCAAAGGAAAUGAGAAAAAUUUUCUUCAUUUCCGUUAAAUGUCACUACACUUUGACUCCGG ........................(((((((((((.....))).))))))))(((((((((....))).))))))..... (-12.62 = -13.32 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:35 2011