
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,886,735 – 19,886,842 |
| Length | 107 |
| Max. P | 0.999662 |

| Location | 19,886,735 – 19,886,842 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 73.21 |
| Shannon entropy | 0.50880 |
| G+C content | 0.43702 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.19 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.997621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19886735 107 + 22422827 ACUUGCUCGUGCGCUAACUGAUUAAAGUUAAUAUGCACUCAUAUUGGAUUAUAAGUGCUGUCAAAUGGGUUCUUCCCUGUUCUGU-UCCGUUUCGAUCCGCACUGUAG------------- ...(((..(((((.(((((......)))))....(((((.(((......))).))))).((((((((((................-))))))).))).))))).))).------------- ( -23.79, z-score = -1.37, R) >droSim1.chrX 15302549 107 + 17042790 ACUUGCUCGUGCGCUAACUGAUUAAAGUUAAUAUGCACUCAUAUUGGAUUAUGAGUGCUGUCAAAUGGGUUCUGCCCUGUUCUGU-UCCGUUUCGAUCCGCACUGGAG------------- .....((.(((((.(((((......)))))....(((((((((......))))))))).((((((((((................-))))))).))).))))).))..------------- ( -29.99, z-score = -2.13, R) >droSec1.super_8 2117754 107 + 3762037 ACUUGCUCGUGCGCUAACUGAUUAAAGUUAAUAUGCACUCAUAUUGGAUUAUGAGUGCUGUCAAAUGGGUUCUGCCCUGUUCUGU-UCCGUUUCGAUCCGCACUGGAG------------- .....((.(((((.(((((......)))))....(((((((((......))))))))).((((((((((................-))))))).))).))))).))..------------- ( -29.99, z-score = -2.13, R) >droYak2.chrX 10233868 90 - 21770863 ACUUGCUCGUGCGCUAACUGAUUAAAGUUAAUAUGCACUCAUAUUGGAUUAUGAGUGCUGUCAAAUGGGUUCUGUUCC--CCCGU-UCUGUUC---------------------------- ....((....))..(((((......)))))....(((((((((......))))))))).....((((((.........--)))))-)......---------------------------- ( -22.80, z-score = -2.33, R) >dp4.chrXL_group1e 7332204 102 + 12523060 GCUUACUCGUGCGCUAACUGAUUAAAGUUAAUAUGCACUCAUAUUGGAUUAUGAGUGCUGUCAAAUGGGUUUCGCCCAUGUGUGUGUGCACGUGUACGAGUG------------------- ...((((((((((((((((......)))))....(((((((((......)))))))))......(((((.....)))))((((....)))))))))))))))------------------- ( -42.50, z-score = -5.33, R) >droVir3.scaffold_12970 10030647 81 + 11907090 ACUUACUCGUGCGCCAACUGAUUAAAGUUAAUAUGCACUCAUAUUGGAUUAUGAGUGCUGUCAAAUGGGUCUAAGCGGAAC---------------------------------------- ......((((..(((((((......)))).....(((((((((......))))))))).........)))....))))...---------------------------------------- ( -21.70, z-score = -2.16, R) >droGri2.scaffold_14853 7312160 119 - 10151454 ACUUACUCGUGCGCCAACUGAUUAAAGUUAAUAUGCACUCAUAUUGGAUUAUGAGUGCUGUCAAAUGGGUCUG-GCAGGCUGUGGAUGUGGGU-UGAGGGUGUGGUUAGGCUGUGUGGUGU ..........(((((((((......)))).(((((((((((((......))))))))).........((((((-(((.(((.(.(((....))-).).))).).))))))))))))))))) ( -36.20, z-score = -1.24, R) >consensus ACUUGCUCGUGCGCUAACUGAUUAAAGUUAAUAUGCACUCAUAUUGGAUUAUGAGUGCUGUCAAAUGGGUUCUGCCCUGUUCUGU_UCCGUUU_GAUCCGCA__G________________ ....((((((..(((((((......)))))....(((((((((......))))))))).))...))))))................................................... (-20.00 = -20.19 + 0.18)

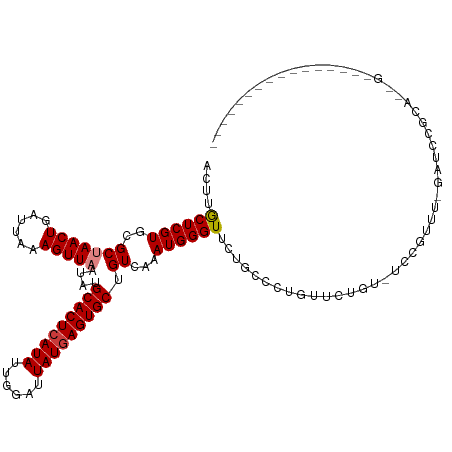
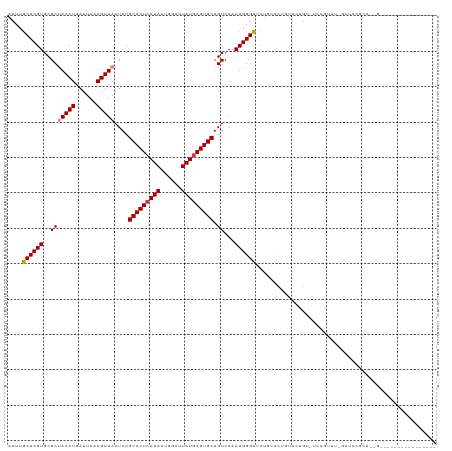
| Location | 19,886,735 – 19,886,842 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 73.21 |
| Shannon entropy | 0.50880 |
| G+C content | 0.43702 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -16.51 |
| Energy contribution | -15.94 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.15 |
| SVM RNA-class probability | 0.999662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19886735 107 - 22422827 -------------CUACAGUGCGGAUCGAAACGGA-ACAGAACAGGGAAGAACCCAUUUGACAGCACUUAUAAUCCAAUAUGAGUGCAUAUUAACUUUAAUCAGUUAGCGCACGAGCAAGU -------------.....(((((..(((((..(..-......).(((.....))).)))))..(((((((((......)))))))))...((((((......)))))))))))........ ( -27.00, z-score = -3.08, R) >droSim1.chrX 15302549 107 - 17042790 -------------CUCCAGUGCGGAUCGAAACGGA-ACAGAACAGGGCAGAACCCAUUUGACAGCACUCAUAAUCCAAUAUGAGUGCAUAUUAACUUUAAUCAGUUAGCGCACGAGCAAGU -------------(((..(((((..(((((..(..-......).(((.....))).)))))..(((((((((......)))))))))...((((((......))))))))))))))..... ( -29.40, z-score = -3.11, R) >droSec1.super_8 2117754 107 - 3762037 -------------CUCCAGUGCGGAUCGAAACGGA-ACAGAACAGGGCAGAACCCAUUUGACAGCACUCAUAAUCCAAUAUGAGUGCAUAUUAACUUUAAUCAGUUAGCGCACGAGCAAGU -------------(((..(((((..(((((..(..-......).(((.....))).)))))..(((((((((......)))))))))...((((((......))))))))))))))..... ( -29.40, z-score = -3.11, R) >droYak2.chrX 10233868 90 + 21770863 ----------------------------GAACAGA-ACGGG--GGAACAGAACCCAUUUGACAGCACUCAUAAUCCAAUAUGAGUGCAUAUUAACUUUAAUCAGUUAGCGCACGAGCAAGU ----------------------------...((((-...((--(........))).))))...(((((((((......)))))))))....(((((......)))))((......)).... ( -24.00, z-score = -3.83, R) >dp4.chrXL_group1e 7332204 102 - 12523060 -------------------CACUCGUACACGUGCACACACACAUGGGCGAAACCCAUUUGACAGCACUCAUAAUCCAAUAUGAGUGCAUAUUAACUUUAAUCAGUUAGCGCACGAGUAAGC -------------------.((((((....((((.....((.(((((.....))))).))...(((((((((......)))))))))....(((((......))))))))))))))).... ( -34.40, z-score = -5.63, R) >droVir3.scaffold_12970 10030647 81 - 11907090 ----------------------------------------GUUCCGCUUAGACCCAUUUGACAGCACUCAUAAUCCAAUAUGAGUGCAUAUUAACUUUAAUCAGUUGGCGCACGAGUAAGU ----------------------------------------((..(((................(((((((((......)))))))))....(((((......)))))))).))........ ( -18.50, z-score = -2.16, R) >droGri2.scaffold_14853 7312160 119 + 10151454 ACACCACACAGCCUAACCACACCCUCA-ACCCACAUCCACAGCCUGC-CAGACCCAUUUGACAGCACUCAUAAUCCAAUAUGAGUGCAUAUUAACUUUAAUCAGUUGGCGCACGAGUAAGU ..........(((((((..........-................((.-((((....)))).))(((((((((......)))))))))................))))).)).......... ( -21.50, z-score = -2.46, R) >consensus ________________C__UGCGGAUC_AAACGGA_ACAGAACAGGGCAGAACCCAUUUGACAGCACUCAUAAUCCAAUAUGAGUGCAUAUUAACUUUAAUCAGUUAGCGCACGAGCAAGU ...............................................................(((((((((......)))))))))....(((((......)))))((......)).... (-16.51 = -15.94 + -0.57)
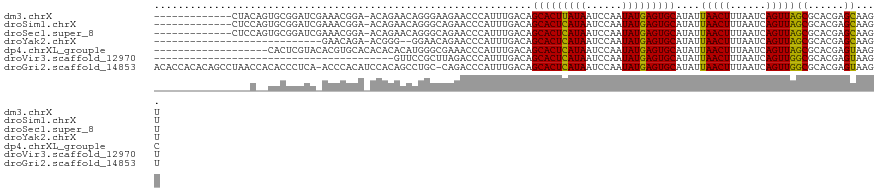
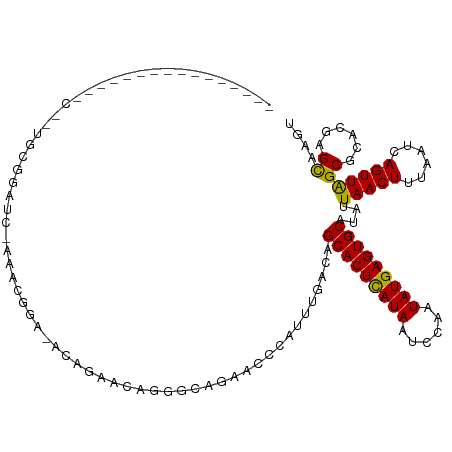
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:31 2011