| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,090,798 – 1,090,897 |
| Length | 99 |
| Max. P | 0.536375 |

| Location | 1,090,798 – 1,090,897 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.16 |
| Shannon entropy | 0.61631 |
| G+C content | 0.39911 |
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -8.51 |
| Energy contribution | -8.71 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1090798 99 + 23011544 ------UUACACUCAUUUUUGGGUCAAGUUCAAUUGCAGGCAUUAAUUAUAA-GGACUUGGCACUUGGACUUGUUCGCC-UUCGAGU-UCAAGUGAAUGCAACUCGCA- ------....(((((....)))))(((((((.....................-))))))).((((((((((((......-..)))))-)))))))..(((.....)))- ( -26.60, z-score = -2.00, R) >droPer1.super_8 717254 87 + 3966273 --------UCACACAUUUUUGGGUCAAGUUCAAUUGCAAACAUUAAUUAUAA-GGACUUGGCACUCU---UGGCGCUCAAAACGAAU-UCGAA-AAGUGCA-------- --------...(((..(((((((((((((((.....................-)))))))((.(...---.)))))))))))((...-.))..-..)))..-------- ( -16.10, z-score = -0.83, R) >dp4.chr4_group3 9529779 87 + 11692001 --------UCACACAUUUUUGGGUCAAGUUCAAUUGCAAACAUUAAUUAUAA-GGACUUGGCACUCU---UGGCGCUCAAAACAAAU-UCGAA-AAGUGCA-------- --------...(((.((((((((((((((((.....................-)))))))))....(---((..........)))..-)))))-)))))..-------- ( -15.60, z-score = -0.99, R) >droAna3.scaffold_12916 4556450 84 + 16180835 ----UCUCUUUCUAUUUUUGGGGUCAAGCUCAACUGCAGGCAUUAAUUAUAA-GGACUUGGCACUUG-----UUUGGCUUCUCAAGU-UCAAAUG-------------- ----....(((..((((..((((((((((.....(((((((.(((....)))-.).))).)))...)-----)))))))))..))))-..)))..-------------- ( -14.60, z-score = 0.07, R) >droEre2.scaffold_4929 1135590 97 + 26641161 ------UUGCACACAUUUUUGGGUCAAGUUCAAUUGCAGGCAUUAAUUAUAA-GGACUUGGCACUCU--CUCGUUCGCCACUCGAGU-UCAAAUGAACUCAACUUGC-- ------..(((........((((((((((((.....................-)))))))))((...--...))...)))...((((-((....))))))....)))-- ( -19.90, z-score = -0.77, R) >droYak2.chr2L 1067484 98 + 22324452 ------UUGCACACAUUUUUGGGUCAAGCUCAAUUGCAGGCAUUAAUUAUAA-GGACUUGGCACUCU--CUUGUUCGCCACUUGAGU-UCAAAUGAAUUCAACUUGCA- ------............(((((.....))))).((((((............-.....((((((...--...))..)))).((((((-((....))))))))))))))- ( -21.30, z-score = -0.64, R) >droSec1.super_14 1044159 97 + 2068291 ------UUACACUCAUUUUUGGGUCAAGUUCAAUUGCAGGCAUUAAUUAUAA-GGACUUGGCACUCU--CUUGUUCGCC-CUCGAGU-UCAAGUGAAUUCAACUCGAA- ------...........(((((((..((((((..((....))..........-(((((((((((...--...))..)))-...))))-))...))))))..)))))))- ( -18.50, z-score = -0.18, R) >droSim1.chr2L 1061514 92 + 22036055 ------UUACACUCAUUUUUGGGUCAAGUUCAAUUGCAGGCAUUAAUUAUAA-GGACUUGGCACUCU--CCUGUUCGCC-CUCGAGU-UCAAGUGAAUUCGCA------ ------....(((((....)))))..........(((.(((.(((....)))-((((..((......--)).)))))))-...((((-((....)))))))))------ ( -18.50, z-score = -0.24, R) >droMoj3.scaffold_6500 22999769 90 + 32352404 UUAUUCUCAUUC-GCAUUUUAGGUCAAGUUCAAUUGGCAACAUUAAUUCUAAAGGACUUGGCACUUG-----GAUG-CGGCACAAGU-UCAAAUAAAU----------- ..........((-(((((..(((((((((((...((....))(((....))).))))))))).))..-----))))-))).......-..........----------- ( -23.50, z-score = -4.47, R) >droWil1.scaffold_180772 4688132 92 - 8906247 ---------------UUUUUGGGUCAAGUUCAAUUGC-AACAUUAAUUAUAA-GGACUUGGCACUUGUUAUCGCCUCCAAUCUGAACCUCCCCCGCACUUGCCACACUU ---------------.....(((..(.(((((((((.-.....)))))....-(((...(((..........)))))).....)))).)..)))((....))....... ( -14.40, z-score = 0.29, R) >consensus ______UUACACACAUUUUUGGGUCAAGUUCAAUUGCAGGCAUUAAUUAUAA_GGACUUGGCACUCU__CUUGUUCGCCACUCGAGU_UCAAAUGAAUUCA_C______ ......................(((((((((......................)))))))))............................................... ( -8.51 = -8.71 + 0.20)

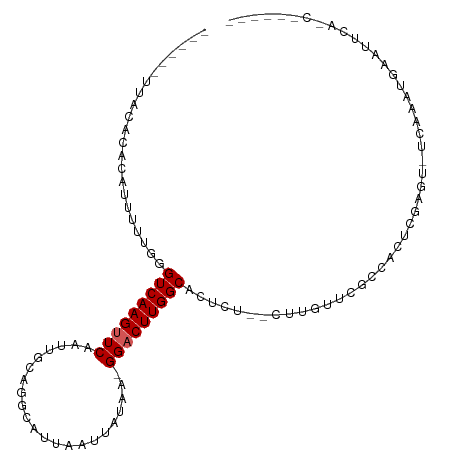
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:42 2011