| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,826,406 – 19,826,513 |
| Length | 107 |
| Max. P | 0.823825 |

| Location | 19,826,406 – 19,826,513 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.01 |
| Shannon entropy | 0.27987 |
| G+C content | 0.44041 |
| Mean single sequence MFE | -22.79 |
| Consensus MFE | -11.60 |
| Energy contribution | -12.72 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.583604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19826406 107 + 22422827 UCCUCGAUUUCCC-UGGCAGGUUCUUAUCAUCGUAGCAGCAAUACCACUGCUAUUACUAGUACUAUCUAGUGGCACUACCUUCGAUCCAUACCAAAUCAAGUUCACAC .....(((((...-(((.((((..........(((((((........)))))))((((((......)))))).....)))).....)))....))))).......... ( -19.80, z-score = -1.64, R) >droSim1.chrX 15266052 108 + 17042790 UCCUCGAUUUCCCCUGGCAGGUUUUCAUCAUCGUAGCAGCAAAACCACUGCUAUUAUUGCUACUACCUAGUGGUACUACCUUCGAUCCAGACCAAAUCAAGUUCACAC .....(((((...((((.((((..........(((((((........)))))))...(((((((....)))))))..)))).....))))...))))).......... ( -24.00, z-score = -2.65, R) >droSec1.super_2418 3990 108 + 4662 UCCUCGAUUUCCCCUGGCAGGUUUUCAUCAUCGUAGCAGCAAAACCACUGCUAUUAUUGCUACUACCUAGUGGUACUACCUUCGAUCCAGACCAAAUCGAGUUCACAC ..((((((((...((((.((((..........(((((((........)))))))...(((((((....)))))))..)))).....))))...))))))))....... ( -29.80, z-score = -4.24, R) >droYak2.chrX 10198328 104 - 21770863 -UCUCCAUUUUCCCUUAAAGAUUUUCAUCAUCGUAGCAGCACAACUACUGUUAUUGCUGGGAUUACCUAGU---GCUACCUUCGAUCCAGACCAAAUCAAGUUCGCAC -..................(((((((...((((.((.(((..(((....)))...((((((....))))))---)))..)).))))...))..))))).......... ( -16.50, z-score = -0.61, R) >droEre2.scaffold_4690 10004363 104 + 18748788 UCCCCGAUUUCCC-UGGAAGGUUUUCAUCGUUGCAACAGCAAAACCAGUGUUAUUACUGGUUUUACCUACU---ACUAUCUUCGAUCCAGACCAAAUCAAGUUCACAC .....(((((..(-((((((((.......((((...))))((((((((((....))))))))))))))...---...........)))))...))))).......... ( -23.83, z-score = -2.90, R) >consensus UCCUCGAUUUCCCCUGGCAGGUUUUCAUCAUCGUAGCAGCAAAACCACUGCUAUUACUGGUACUACCUAGUGG_ACUACCUUCGAUCCAGACCAAAUCAAGUUCACAC ...................(((((..(((...(((((((........)))))))((((((......))))))...........)))..)))))............... (-11.60 = -12.72 + 1.12)

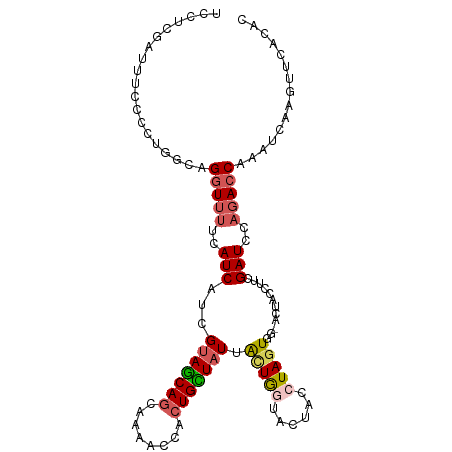
| Location | 19,826,406 – 19,826,513 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.01 |
| Shannon entropy | 0.27987 |
| G+C content | 0.44041 |
| Mean single sequence MFE | -31.39 |
| Consensus MFE | -20.58 |
| Energy contribution | -21.46 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19826406 107 - 22422827 GUGUGAACUUGAUUUGGUAUGGAUCGAAGGUAGUGCCACUAGAUAGUACUAGUAAUAGCAGUGGUAUUGCUGCUACGAUGAUAAGAACCUGCCA-GGGAAAUCGAGGA .......((((((((((((.((((((..((((((((((((.(....((....))....)))))))))))))....))))........)))))))-....))))))).. ( -30.50, z-score = -2.00, R) >droSim1.chrX 15266052 108 - 17042790 GUGUGAACUUGAUUUGGUCUGGAUCGAAGGUAGUACCACUAGGUAGUAGCAAUAAUAGCAGUGGUUUUGCUGCUACGAUGAUGAAAACCUGCCAGGGGAAAUCGAGGA .......((((((((..(((((((((..(.((.((((....)))).)).).....(((((((......)))))))))))..(....)....)))))..)))))))).. ( -33.70, z-score = -2.40, R) >droSec1.super_2418 3990 108 - 4662 GUGUGAACUCGAUUUGGUCUGGAUCGAAGGUAGUACCACUAGGUAGUAGCAAUAAUAGCAGUGGUUUUGCUGCUACGAUGAUGAAAACCUGCCAGGGGAAAUCGAGGA .......((((((((..(((((((((..(.((.((((....)))).)).).....(((((((......)))))))))))..(....)....)))))..)))))))).. ( -35.80, z-score = -2.86, R) >droYak2.chrX 10198328 104 + 21770863 GUGCGAACUUGAUUUGGUCUGGAUCGAAGGUAGC---ACUAGGUAAUCCCAGCAAUAACAGUAGUUGUGCUGCUACGAUGAUGAAAAUCUUUAAGGGAAAAUGGAGA- ((((...((((((((.....))))).)))...))---)).......((((((((.((((....))))))))........(((....))).....)))).........- ( -25.40, z-score = -0.90, R) >droEre2.scaffold_4690 10004363 104 - 18748788 GUGUGAACUUGAUUUGGUCUGGAUCGAAGAUAGU---AGUAGGUAAAACCAGUAAUAACACUGGUUUUGCUGUUGCAACGAUGAAAACCUUCCA-GGGAAAUCGGGGA .......((((((((..((((((.........((---.(((((((((((((((......))))))))))))..))).))...........))))-)).)))))))).. ( -31.55, z-score = -2.56, R) >consensus GUGUGAACUUGAUUUGGUCUGGAUCGAAGGUAGU_CCACUAGGUAGUACCAGUAAUAGCAGUGGUUUUGCUGCUACGAUGAUGAAAACCUGCCAGGGGAAAUCGAGGA .......((((((((..(((((((((..((((((((((((.(.((.((....)).)).)))))))...))))))....)))).........)))))..)))))))).. (-20.58 = -21.46 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:25 2011