
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,809,554 – 19,809,656 |
| Length | 102 |
| Max. P | 0.845586 |

| Location | 19,809,554 – 19,809,656 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.74 |
| Shannon entropy | 0.72200 |
| G+C content | 0.63122 |
| Mean single sequence MFE | -45.09 |
| Consensus MFE | -19.44 |
| Energy contribution | -18.84 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19809554 102 + 22422827 UUCUGGUAACCAUAG---GUACCCGGGUAGUUGGGG------------CCCAU---GUAGCCCGGCUGGCCGGGAUGGCCCACCUGGUUGUUGUAGGAGAAGGGACGACCGUGCUCGUCC (((((.((((((..(---((.(((((.((((((((.------------.....---....)))))))).)))))...)))....))))))...)))))....((((((......)))))) ( -41.20, z-score = 0.07, R) >droSec1.super_8 2075798 114 + 3762037 UUCUGGUAGCCAUAG---GCACCCGGGUAGUUGGGGUCCGGCCGUACGCCCAU---GUAGCCCGGCUGGCCGGGCUGGCCCACCUGGUUGCUGUAGGAGAAGGGGCGACCGUGCUCGUCU ....((((((((..(---((.(((((.((((((((.((.(((.....)))...---).).)))))))).)))))...)))....))))))))....(((.(.((....)).).))).... ( -49.90, z-score = 1.05, R) >droYak2.chrX 10201080 114 - 21770863 UUGUGGUAGCCAUAG---GAUCCCGGGUAGUUGGGAUCCGGCCGUACGCCCAU---GUAGCCCGGCUGACCCGGCUGACCCACCUGGUUGCCGUAGGAGAAGGGACGACCAUGCUCGUCA ((((((((((((..(---(.(((((((((((((((....(((.....)))...---....))))))).))))))..)).))...)))))))))))).......(((((......))))). ( -46.10, z-score = -0.36, R) >droEre2.scaffold_4690 10006449 114 + 18748788 UUCUGGUAGCCAUAG---GAUCCCGGGUAGCUGGGAUCCGGCCGUACGCCCAU---GUAGCCCGGCUGUCCGGGCUGGCCCACCUGGUUGCCGUAGGAGAAGGGGCGACCGUGCUCGUCA ....((((((((..(---((((((((....)))))))))((((.((((....)---)))((((((....)))))).))))....))))))))....(((.(.((....)).).))).... ( -57.40, z-score = -1.61, R) >droAna3.scaffold_13417 4997255 105 - 6960332 UUCCGGUAGCCGUAGACUCCGCCCGGAUAGUUGGGAUC---CCGGACACCCAU---GUAGCCCGGCUGUCCAGGC---------UGGCUGCCGUACGAGAACGGACGUCCGUGCUCGUCC ...(((((((((....)...(((.(((((((((((...---..((....))..---....))))))))))).)))---------.)))))))).(((((.((((....)))).))))).. ( -52.20, z-score = -3.50, R) >dp4.chrXL_group1e 5152404 114 - 12523060 CCCCGAUAGCCAUAG---CUGCCGGGAUAUCCCGGAUCGAGUCGGCCUCCCAUAUUGUAGCCCGGCUGUCCGGGCUGUCCC---UGUCCGUUGUAGGAGAAGGGUCGUCCGUGCUCAUCC .((((.((((....)---))).)))).......((((.((((((((..(((......((((((((....))))))))....---(.(((......))).).)))..).))).)))))))) ( -47.90, z-score = -2.33, R) >droPer1.super_13 1177852 114 + 2293547 CCCCGAUAGCCAUAG---CUGCCGGGAUAUCCCGGAUCGAGUCGGCCUCCCAUGUUGUAGCCCGGCUGUCCGGGCUGCCCC---UGUCCGUUGUAGGAGAAGGGCCGUCCGUGCUCAUCC .((((.((((....)---))).)))).......((((.(((((((.(.........(((((((((....)))))))))(((---(.(((......)))..))))..).))).)))))))) ( -50.10, z-score = -2.49, R) >droWil1.scaffold_180777 3872730 93 - 4753960 CCACGAUAACCAUAA---UAAUCAUUACGCCCGGGA----------UAGCCAGGCGGUGGAAU--UUGGCCA--CC----------ACCAUUAUAGGAGAAGGGUCGCCCAUGCUCAUCC (((((((........---..)))....((((.((..----------...)).))))))))...--..(((.(--((----------.((......)).....))).)))........... ( -23.30, z-score = 0.40, R) >droVir3.scaffold_12970 1825745 94 - 11907090 --UCUAUAUCCAUAG---UUGUUCGGAUAGCCGGG-----------CGGUGGGGGCACGGGCUGGCCGCCCAUGCC----------GCCAUUAUAGGAAUAGGGUCGUCCAUGCUCGUCG --(((((.(((....---..((((((....)))))-----------)(((((.((((.((((.....)))).))))----------.)))))...))))))))(.((........)).). ( -37.70, z-score = -0.68, R) >consensus UUCCGGUAGCCAUAG___GUGCCCGGAUAGCUGGGAUC__GCCG_ACGCCCAU___GUAGCCCGGCUGUCCGGGCUG_CCC___UGGCUGUUGUAGGAGAAGGGACGACCGUGCUCGUCC ...((((..............(((((....))))).......................(((((((....))))))).............))))...(((...((....))...))).... (-19.44 = -18.84 + -0.60)

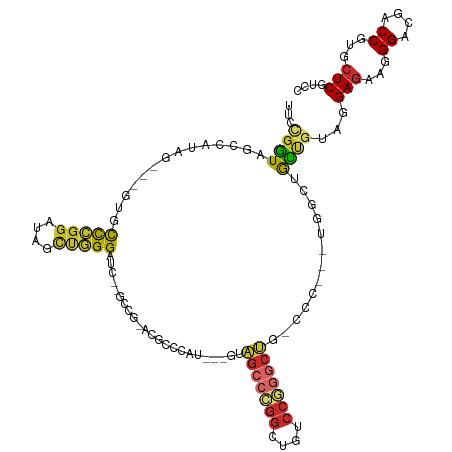
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:24 2011