
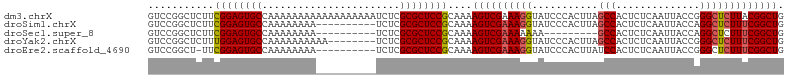
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,778,751 – 19,778,858 |
| Length | 107 |
| Max. P | 0.952035 |

| Location | 19,778,751 – 19,778,857 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 88.67 |
| Shannon entropy | 0.19094 |
| G+C content | 0.50674 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.56 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
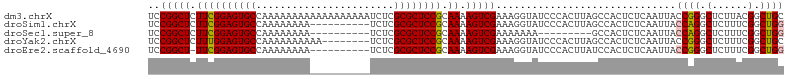
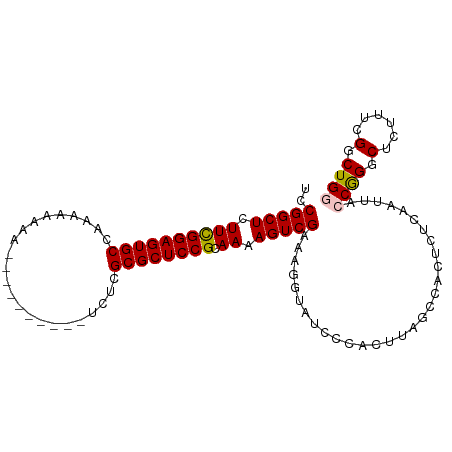
>dm3.chrX 19778751 106 + 22422827 GUCCGGCUCUUCGGAGUGCCAAAAAAAAAAAAAAAAAAUCUCGCGCUCCGCAAAAGUCGAAAGGUAUCCCACUUAGCCACUCUCAAUUACCGGGCUCUUACGGCUG ((((((((.((((((((((.......................)))))))).)).))))....((((.....................))))))))........... ( -25.30, z-score = -1.87, R) >droSim1.chrX 15249328 96 + 17042790 GUCCGGCUCUUCGGAGUGCCAAAAAAAA----------UCUCGCGCUCCGCAAAAGUCGAAAGGUAUCCCACUUAGCCACUCUCAAUUACCAGGCUCUUUCGGCUG (((.((((.((((((((((.........----------....)))))))).)).))))(((((((.....))..((((..............)))))))))))).. ( -25.66, z-score = -2.23, R) >droSec1.super_8 2042061 87 + 3762037 GUCCGGCUCUUCGGAGUGCCAAAAAAAA----------UCUCGCGCUCCGCAAAAGUCGAAAAAAA---------GCCACUCUCAAUUACCAGGCUCUUUCGGCUG (((.((((.((((((((((.........----------....)))))))).)).))))((((...(---------(((..............)))).))))))).. ( -24.76, z-score = -3.00, R) >droYak2.chrX 10175266 98 - 21770863 GUCCGGCUCUUUGGAGUGCCAAAAAAAAAA--------UCUCGCGCUCCGCAAAAGUCGAAAGGUAUCCCACUUAGCCACUCUCAAUUACCGGGCUCUUUCGGCUG ............(((((((...........--------....))))))).....((((((((((...(((.....................))).)))))))))). ( -24.96, z-score = -1.40, R) >droEre2.scaffold_4690 9981941 95 + 18748788 GUCCGGCU-UUCGGAGUGCCAAAAAAAA----------UCUCGCGCUCCGCAAAAGUCGAAAGGUAUCCCACUUAUCCACUCUCAAUUACCGGGCUCUUUCGGCUG (((.((((-((((((((((.........----------....))))))))..))))))((((((...(((.....................))).))))))))).. ( -27.12, z-score = -2.80, R) >consensus GUCCGGCUCUUCGGAGUGCCAAAAAAAA__________UCUCGCGCUCCGCAAAAGUCGAAAGGUAUCCCACUUAGCCACUCUCAAUUACCGGGCUCUUUCGGCUG ((((((((.((((((((((.......................)))))))).)).)))))................(((..............)))......))).. (-21.52 = -21.56 + 0.04)

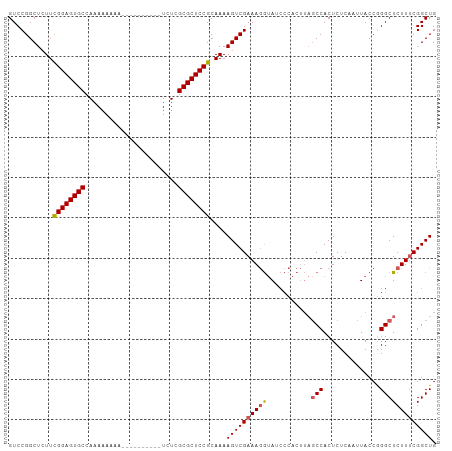
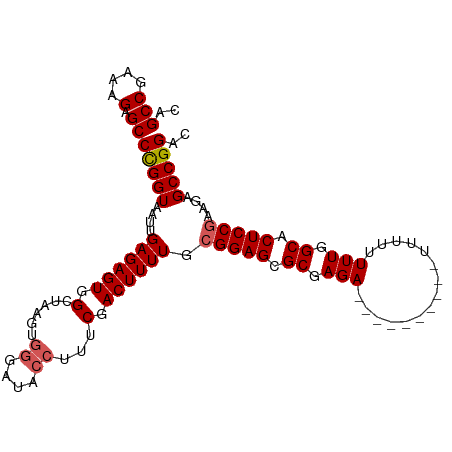
| Location | 19,778,751 – 19,778,857 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Shannon entropy | 0.19094 |
| G+C content | 0.50674 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -25.84 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19778751 106 - 22422827 CAGCCGUAAGAGCCCGGUAAUUGAGAGUGGCUAAGUGGGAUACCUUUCGACUUUUGCGGAGCGCGAGAUUUUUUUUUUUUUUUUUUGGCACUCCGAAGAGCCGGAC ..(((....).))((((...(((((((.(.((.....))...))))))))(((((.(((((.((.(((...............))).)).)))))))))))))).. ( -30.06, z-score = -0.66, R) >droSim1.chrX 15249328 96 - 17042790 CAGCCGAAAGAGCCUGGUAAUUGAGAGUGGCUAAGUGGGAUACCUUUCGACUUUUGCGGAGCGCGAGA----------UUUUUUUUGGCACUCCGAAGAGCCGGAC ..(((....).))((((...(((((((.(.((.....))...))))))))(((((.(((((.((.(((----------.....))).)).)))))))))))))).. ( -30.20, z-score = -1.09, R) >droSec1.super_8 2042061 87 - 3762037 CAGCCGAAAGAGCCUGGUAAUUGAGAGUGGC---------UUUUUUUCGACUUUUGCGGAGCGCGAGA----------UUUUUUUUGGCACUCCGAAGAGCCGGAC .....(((((((((..............)))---------))))))(((.(((((.(((((.((.(((----------.....))).)).)))))))))).))).. ( -28.74, z-score = -1.47, R) >droYak2.chrX 10175266 98 + 21770863 CAGCCGAAAGAGCCCGGUAAUUGAGAGUGGCUAAGUGGGAUACCUUUCGACUUUUGCGGAGCGCGAGA--------UUUUUUUUUUGGCACUCCAAAGAGCCGGAC ..(((....).))(((((...........((.((((.(((.....))).))))..))((((.((.(((--------.......))).)).)))).....))))).. ( -31.60, z-score = -1.51, R) >droEre2.scaffold_4690 9981941 95 - 18748788 CAGCCGAAAGAGCCCGGUAAUUGAGAGUGGAUAAGUGGGAUACCUUUCGACUUUUGCGGAGCGCGAGA----------UUUUUUUUGGCACUCCGAA-AGCCGGAC ..(((....).))(((((....((((((.((.....((....))..)).)))))).(((((.((.(((----------.....))).)).)))))..-.))))).. ( -34.40, z-score = -2.96, R) >consensus CAGCCGAAAGAGCCCGGUAAUUGAGAGUGGCUAAGUGGGAUACCUUUCGACUUUUGCGGAGCGCGAGA__________UUUUUUUUGGCACUCCGAAGAGCCGGAC ..(((....).))(((((....((((((.(......((....))...).)))))).(((((.((.(((...............))).)).)))))....))))).. (-25.84 = -26.20 + 0.36)

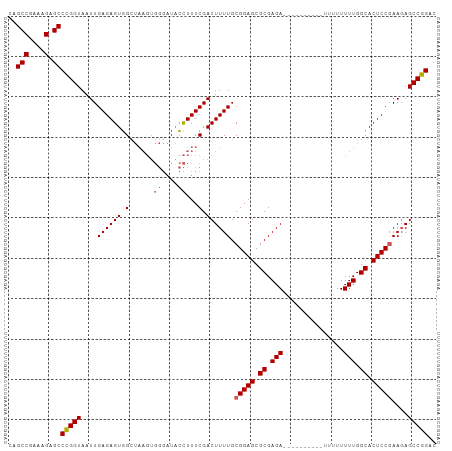
| Location | 19,778,752 – 19,778,858 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 88.07 |
| Shannon entropy | 0.20010 |
| G+C content | 0.50674 |
| Mean single sequence MFE | -26.29 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19778752 106 + 22422827 UCCGGCUCUUCGGAGUGCCAAAAAAAAAAAAAAAAAAUCUCGCGCUCCGCAAAAGUCGAAAGGUAUCCCACUUAGCCACUCUCAAUUACCGGGCUCUUACGGCUGC ..(((((.((((((((((.......................)))))))).)).)))))....(((.((.....((((..............)))).....)).))) ( -24.54, z-score = -1.19, R) >droSim1.chrX 15249329 96 + 17042790 UCCGGCUCUUCGGAGUGCCAAAAAAAA----------UCUCGCGCUCCGCAAAAGUCGAAAGGUAUCCCACUUAGCCACUCUCAAUUACCAGGCUCUUUCGGCUGG .(((((....((((((((.........----------....)))))))).......((((((((.....))..((((..............))))))))))))))) ( -27.26, z-score = -2.31, R) >droSec1.super_8 2042062 87 + 3762037 UCCGGCUCUUCGGAGUGCCAAAAAAAA----------UCUCGCGCUCCGCAAAAGUCGAAAAAAA---------GCCACUCUCAAUUACCAGGCUCUUUCGGCUGG .(((((....((((((((.........----------....)))))))).......(((((...(---------(((..............)))).)))))))))) ( -26.36, z-score = -3.12, R) >droYak2.chrX 10175267 98 - 21770863 UCCGGCUCUUUGGAGUGCCAAAAAAAAAA--------UCUCGCGCUCCGCAAAAGUCGAAAGGUAUCCCACUUAGCCACUCUCAAUUACCGGGCUCUUUCGGCUGC ...........(((((((...........--------....))))))).....((((((((((...(((.....................))).)))))))))).. ( -24.96, z-score = -0.97, R) >droEre2.scaffold_4690 9981942 95 + 18748788 UCCGGCU-UUCGGAGUGCCAAAAAAAA----------UCUCGCGCUCCGCAAAAGUCGAAAGGUAUCCCACUUAUCCACUCUCAAUUACCGGGCUCUUUCGGCUGG .((((((-((((((((((.........----------....)))))))).)))...(((((((...(((.....................))).)))))))))))) ( -28.32, z-score = -2.76, R) >consensus UCCGGCUCUUCGGAGUGCCAAAAAAAA__________UCUCGCGCUCCGCAAAAGUCGAAAGGUAUCCCACUUAGCCACUCUCAAUUACCGGGCUCUUUCGGCUGG ..(((((.((((((((((.......................)))))))).)).)))))..............................((((.(......).)))) (-22.24 = -22.24 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:18 2011