| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,775,060 – 19,775,153 |
| Length | 93 |
| Max. P | 0.979203 |

| Location | 19,775,060 – 19,775,153 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 54.19 |
| Shannon entropy | 0.85353 |
| G+C content | 0.47280 |
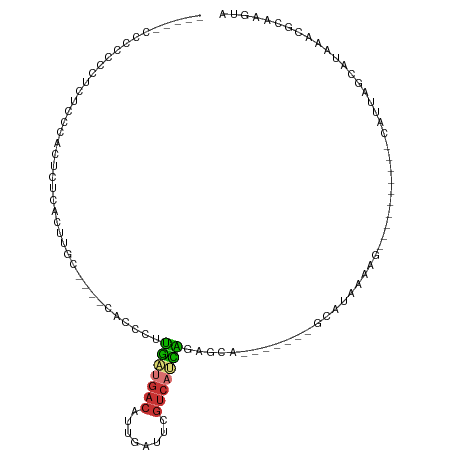
| Mean single sequence MFE | -16.31 |
| Consensus MFE | -5.32 |
| Energy contribution | -5.33 |
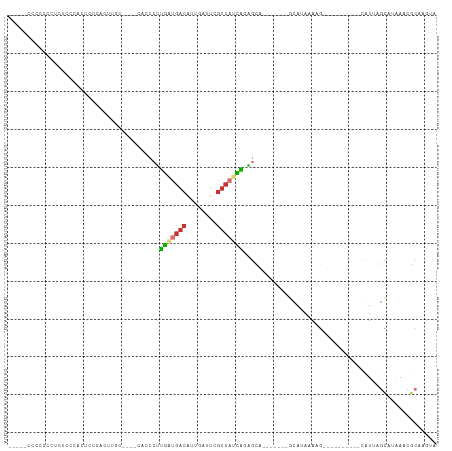
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
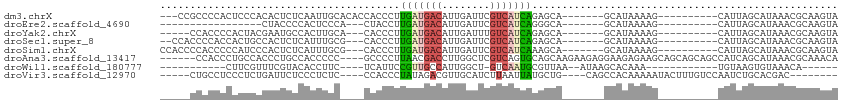
>dm3.chrX 19775060 93 - 22422827 ---CCGCCCCACUCCCACACUCUCAAUUGCACACCACCCUUGAUGACAUUGAUUCGUCAUCAGAGCA-------GCAUAAAAG----------CAUUAGCAUAAACGCAAGUA ---........................(((.........((((((((........))))))))....-------((......)----------)....)))...((....)). ( -16.00, z-score = -2.15, R) >droEre2.scaffold_4690 9978357 76 - 18748788 -----------------CUACCCCACUCCCA---CUACCUUGAUGACAUUGAUUCGUCAUCAGGGCA-------GCAUAAAAG----------CAUUAGCAUAAACGCAAGUA -----------------..............---((.((((((((((........)))))))))).)-------).......(----------(....))....((....)). ( -19.60, z-score = -3.51, R) >droYak2.chrX 10171337 88 + 21770863 -----CCACCCCACUACGAAUGCCACUUGCA---CACCCUUGAUGACAUUGAUUUGUCAUCAGAGCA-------GCAUAAAAG----------CAUUAGCAUAAACGCAAGUA -----...................((((((.---.....(((((((((......))))))))).((.-------((......)----------)....))......)))))). ( -21.20, z-score = -3.58, R) >droSec1.super_8 2038261 91 - 3762037 --CCACCCCACCACUGCCACUCUCAUUUGCG---CACCCUUGAUGACAUUGAUUCGUCAUCAGAGCA-------GCAUAAAAG----------CAUUAGCAUAAACGCAAGUA --......................(((((((---(..(.((((((((........)))))))).)..-------))......(----------(....))......)))))). ( -18.90, z-score = -2.39, R) >droSim1.chrX 15245443 93 - 17042790 CCACCCCACCCCCAUCCCACUCUCAUUUGCG---CACCCUUGAUGACAUUGAUUCGUCAUCAAAGCA-------GCAUAAAAG----------CAUUAGCAUAAACGCAAGUA ........................(((((((---(..(.((((((((........)))))))).)..-------))......(----------(....))......)))))). ( -19.20, z-score = -4.28, R) >droAna3.scaffold_13417 4924512 103 + 6960332 ------CCACCCUGCCACCCUGCCACCCCC----GCCCCUUAACGACCUUGGCUCGUCAGUGCAGCAAGAAGAGGAAGAGAAGCAGCAGCAGCCAUCAGCAUAAACGCAAACA ------.....((((....((((......(----(........)).((((.(((.((....))))).....)))).......))))..))))......((......))..... ( -16.50, z-score = 1.69, R) >droWil1.scaffold_180777 3835249 77 + 4753960 -----------CUUCGUUUCGUACACCUUC----UCAUUCCGUUGCCAUUGGCU-GUCAAUGCGUUAA--AUAAGCACAAA------------UGUAAGUGUAAACA------ -----------....(((...(((((....----.((((.....(((...))).-.....(((.....--....)))..))------------))...)))))))).------ ( -9.30, z-score = 0.81, R) >droVir3.scaffold_12970 1783493 92 + 11907090 -----CUGCCUCCCUCUGAUUCUCCCUCUC----CCACCCUAUAGACGUUGCAUCUUAAUUAUGCUG----CAGCCACAAAAAUACUUUGUCCAAUCUGCACGAC-------- -----.(((.....................----.............((((((.(........).))----)))).(((((.....))))).......)))....-------- ( -9.80, z-score = -0.87, R) >consensus _____CCCCCCCUCUCCCACUCUCACUUGC____CACCCUUGAUGACAUUGAUUCGUCAUCAGAGCA_______GCAUAAAAG__________CAUUAGCAUAAACGCAAGUA ........................................(((((((........)))))))................................................... ( -5.32 = -5.33 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:15 2011