
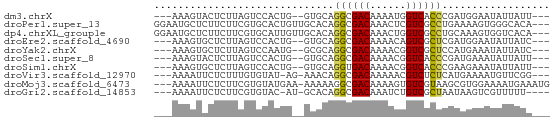
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,770,781 – 19,770,839 |
| Length | 58 |
| Max. P | 0.989252 |

| Location | 19,770,781 – 19,770,839 |
|---|---|
| Length | 58 |
| Sequences | 10 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 62.72 |
| Shannon entropy | 0.75192 |
| G+C content | 0.43905 |
| Mean single sequence MFE | -13.58 |
| Consensus MFE | -7.58 |
| Energy contribution | -7.57 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19770781 58 + 22422827 ---AAAGUACUCUUAGUCCACUG--GUGCAGGCGACAAAAUGGUCACCCGAUGGAAUAUUAUU--- ---...(((((...((....)))--)))).((.(((......))).))...............--- ( -9.20, z-score = 0.63, R) >droPer1.super_13 1139707 63 + 2293547 GGAAUGCUCUUCUUCGUGCACUGUUGCACAGGCGACAAACUCGUCGCCUGAAAAGUGGGCACA--- ....(((((...((((((((....)))))(((((((......))))))))))....)))))..--- ( -25.90, z-score = -3.40, R) >dp4.chrXL_group1e 5114045 63 - 12523060 GGAAUGCUCUUCUUCGUGCAUUGUUGCACAGGCGACAAACUGGUCGCCUGCAAAGUGGUCACA--- ....(((........(((((....)))))(((((((......))))))))))...........--- ( -22.30, z-score = -1.99, R) >droEre2.scaffold_4690 9974438 58 + 18748788 ---AAAGUGCUCUUAGUCCACUG--GUGCAGGCGACAAAACAGUCGCUCGAUGGAAUAUUAUC--- ---....(((.((.((....)))--).)))((((((......))))))...............--- ( -12.50, z-score = -0.47, R) >droYak2.chrX 10167316 58 - 21770863 ---AAAGUGCUCUUAGUCCAAUG--GCGCAGGCGACAAAACGGUCGCUCCAUGAAAUAUUAUC--- ---...(((((.((.....)).)--)))).((((((......))))))...............--- ( -13.60, z-score = -1.21, R) >droSec1.super_8 2033883 58 + 3762037 ---AAAGUACUCUUAGUCCACUG--GUGCAGGCGACAAAACGGUCACCCGAUGAAAUAUUAUU--- ---...(((((...((....)))--)))).((.(((......))).))...............--- ( -9.20, z-score = 0.19, R) >droSim1.chrX 15241157 58 + 17042790 ---AAAGUGCUCUUAGUCCACUG--GUGCAGGUGACAAAACGGUCACCCGAAGAAAUAUUAUU--- ---....(((.((.((....)))--).)))((((((......))))))...............--- ( -13.20, z-score = -1.16, R) >droVir3.scaffold_12970 1779149 58 - 11907090 ---AAAAUUCUCUUUGUGUAU-AG-AAACAGGCGACAAAAACGUCUCUCAUGAAAAUGUUCGG--- ---..................-.(-((..(((((.......)))))..(((....))))))..--- ( -5.90, z-score = 0.54, R) >droMoj3.scaffold_6473 11326292 62 + 16943266 ---AAAAUUCUCUUCGUGUAUGAA-AAAAAGGCGACAAAAGUGUCGUAAGCGUGGAAAAUGAAAUG ---...(((.(((.(((.......-......((((((....))))))..))).))).)))...... ( -10.26, z-score = -1.40, R) >droGri2.scaffold_14853 3607693 57 - 10151454 ---AAAAUUCUCUUCGUGUAC-AU-GCACAGGCGACAAAUCUGUCGCUAAUAAGUCGUUUUU---- ---............((((..-..-)))).(((((((....)))))))..............---- ( -13.70, z-score = -2.87, R) >consensus ___AAAGUGCUCUUAGUCCACUG__GCACAGGCGACAAAACGGUCGCCCGAAGAAAUAUUAUA___ ..............................((((((......)))))).................. ( -7.58 = -7.57 + -0.01)
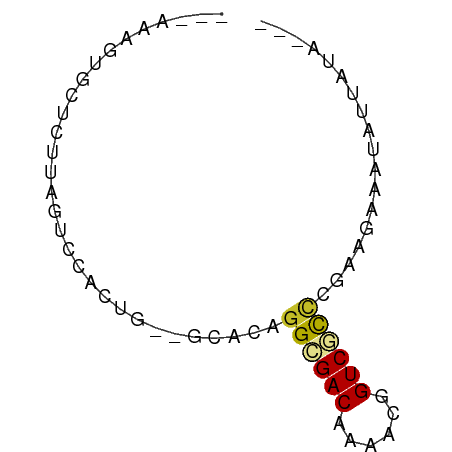

| Location | 19,770,781 – 19,770,839 |
|---|---|
| Length | 58 |
| Sequences | 10 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 62.72 |
| Shannon entropy | 0.75192 |
| G+C content | 0.43905 |
| Mean single sequence MFE | -13.58 |
| Consensus MFE | -5.54 |
| Energy contribution | -5.48 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.952850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19770781 58 - 22422827 ---AAUAAUAUUCCAUCGGGUGACCAUUUUGUCGCCUGCAC--CAGUGGACUAAGAGUACUUU--- ---........(((((((((((((......))))))))...--..))))).............--- ( -15.60, z-score = -2.07, R) >droPer1.super_13 1139707 63 - 2293547 ---UGUGCCCACUUUUCAGGCGACGAGUUUGUCGCCUGUGCAACAGUGCACGAAGAAGAGCAUUCC ---.((((.(..(((((((((((((....))))))))(((((....)))))))))).).))))... ( -24.60, z-score = -3.32, R) >dp4.chrXL_group1e 5114045 63 + 12523060 ---UGUGACCACUUUGCAGGCGACCAGUUUGUCGCCUGUGCAACAAUGCACGAAGAAGAGCAUUCC ---...........((((((((((......)))))))(((((....)))))........))).... ( -21.00, z-score = -2.37, R) >droEre2.scaffold_4690 9974438 58 - 18748788 ---GAUAAUAUUCCAUCGAGCGACUGUUUUGUCGCCUGCAC--CAGUGGACUAAGAGCACUUU--- ---........(((((((.(((((......))))).))...--..))))).............--- ( -12.20, z-score = -0.80, R) >droYak2.chrX 10167316 58 + 21770863 ---GAUAAUAUUUCAUGGAGCGACCGUUUUGUCGCCUGCGC--CAUUGGACUAAGAGCACUUU--- ---........(((((((.(((((......))))).....)--)).)))).............--- ( -10.70, z-score = 0.09, R) >droSec1.super_8 2033883 58 - 3762037 ---AAUAAUAUUUCAUCGGGUGACCGUUUUGUCGCCUGCAC--CAGUGGACUAAGAGUACUUU--- ---........(((((((((((((......))))))))...--..))))).............--- ( -12.70, z-score = -0.77, R) >droSim1.chrX 15241157 58 - 17042790 ---AAUAAUAUUUCUUCGGGUGACCGUUUUGUCACCUGCAC--CAGUGGACUAAGAGCACUUU--- ---.............((((((((......))))))))...--.((((.........))))..--- ( -12.90, z-score = -1.01, R) >droVir3.scaffold_12970 1779149 58 + 11907090 ---CCGAACAUUUUCAUGAGAGACGUUUUUGUCGCCUGUUU-CU-AUACACAAAGAGAAUUUU--- ---......((((((......((((....))))...(((..-..-..)))....))))))...--- ( -6.70, z-score = 0.34, R) >droMoj3.scaffold_6473 11326292 62 - 16943266 CAUUUCAUUUUCCACGCUUACGACACUUUUGUCGCCUUUUU-UUCAUACACGAAGAGAAUUUU--- ....................(((((....)))))..(((((-(((......))))))))....--- ( -8.90, z-score = -3.10, R) >droGri2.scaffold_14853 3607693 57 + 10151454 ----AAAAACGACUUAUUAGCGACAGAUUUGUCGCCUGUGC-AU-GUACACGAAGAGAAUUUU--- ----........(((....(((((......))))).((((.-..-...))))..)))......--- ( -10.50, z-score = -1.01, R) >consensus ___AAUAAUAUUUCAUCGGGCGACCGUUUUGUCGCCUGCAC__CAGUGCACUAAGAGAACUUU___ ...................(((((......)))))............................... ( -5.54 = -5.48 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:14 2011