
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,759,929 – 19,760,061 |
| Length | 132 |
| Max. P | 0.631926 |

| Location | 19,759,929 – 19,760,029 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 67.02 |
| Shannon entropy | 0.50064 |
| G+C content | 0.44856 |
| Mean single sequence MFE | -17.86 |
| Consensus MFE | -9.80 |
| Energy contribution | -10.93 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.631926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
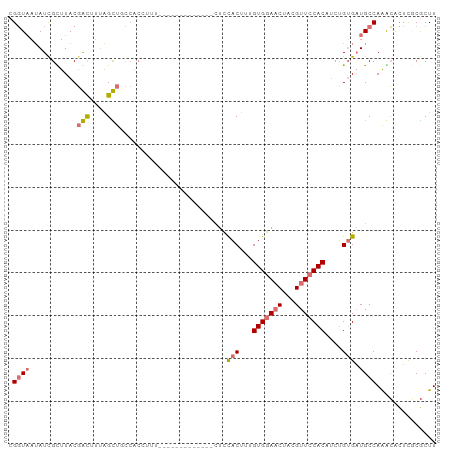
>dm3.chrX 19759929 100 + 22422827 CGGUAAUAUUGUUUACGACAUUAGUUGCCACCUUUACUUAUUUACUUACUUUACUUUGUGGAACUAAGUUCCACAUCUGGGAUGCCAAACACUCACGCUU .((((((..((((...))))...))))))...........................((((((((...))))))))..((((.((.....))))))..... ( -19.10, z-score = -1.64, R) >droSim1.chrX 15234065 87 + 17042790 CGGUAAUAUCGUUUACGGCUUUAGCUUCCACAUUU-------------CUCCACUUUGUGGAACUACGUUCCACAUCUGUGCUGCCAAGCACUCGCGCUU .((((.....((....(((....)))...))....-------------...(((..((((((((...))))))))...))).))))((((......)))) ( -21.50, z-score = -2.16, R) >droSec1.super_8 2023621 87 + 3762037 CGGUAAUAUCGCUUACGGUUUCAGCUUCCACCUUU-------------CUCCACUUUGUGGAACUACGUUACACUUCUGUGAUGCCAAGCACUCGCGCUU .(((...(((((.....((..(((.((((((....-------------.........))))))))..)..))......))))))))((((......)))) ( -16.02, z-score = -0.14, R) >droYak2.chrX 10157936 78 - 21770863 AGCUUAAAUCGCUUACCACUCUUGUUG----------------------UCCACAAAGUGGACCUAUGUUCCACACCAGUGAUGCCAGAGUUUUCCACUU (((.......)))....(((((.((.(----------------------(((((...))))))........(((....)))..)).)))))......... ( -14.80, z-score = -0.83, R) >consensus CGGUAAUAUCGCUUACGACUUUAGCUGCCACCUUU_____________CUCCACUUUGUGGAACUACGUUCCACAUCUGUGAUGCCAAACACUCGCGCUU .((((...........(((....))).........................(((..((((((((...))))))))...))).)))).............. ( -9.80 = -10.93 + 1.12)


| Location | 19,759,949 – 19,760,061 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.95 |
| Shannon entropy | 0.27877 |
| G+C content | 0.46609 |
| Mean single sequence MFE | -23.64 |
| Consensus MFE | -17.22 |
| Energy contribution | -18.67 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
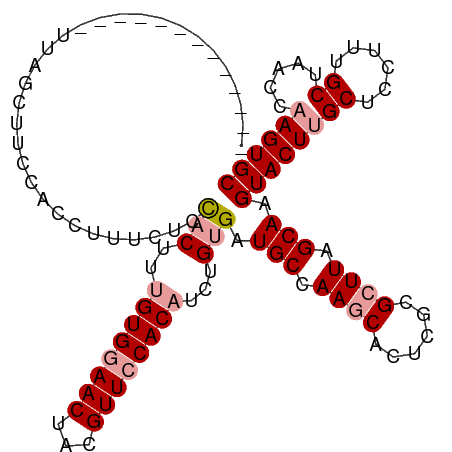
>dm3.chrX 19759949 112 + 22422827 UUAGUUGCCACCUUUACUUAUUUACUUACUUUACUUUGUGGAACUAAGUUCCACAUCUGGGAUGCCAAACACUCACGCUUAGCAAGUACUUGCUCCUCUGCUAACCCAGUGC ....................................((((((((...)))))))).(((((..((...........))(((((((((....)))....)))))))))))... ( -22.00, z-score = -0.77, R) >droSim1.chrX 15234085 99 + 17042790 -------------UUAGCUUCCACAUUUCUCCACUUUGUGGAACUACGUUCCACAUCUGUGCUGCCAAGCACUCGCGCUUAGCAAGUACUUGCUCCUUUGCUAACCAAGUGC -------------(((((..................((((((((...))))))))...(((((((.((((......)))).)).)))))..........)))))........ ( -26.20, z-score = -2.44, R) >droSec1.super_8 2023641 99 + 3762037 -------------UCAGCUUCCACCUUUCUCCACUUUGUGGAACUACGUUACACUUCUGUGAUGCCAAGCACUCGCGCUUAGCAAGUACUUGCUCCUUUGCUAACCAAGUGC -------------..((.((((((.............)))))))).(((((((....)))))))....(((((.(.(.((((((((..........)))))))))).))))) ( -22.72, z-score = -1.32, R) >consensus _____________UUAGCUUCCACCUUUCUCCACUUUGUGGAACUACGUUCCACAUCUGUGAUGCCAAGCACUCGCGCUUAGCAAGUACUUGCUCCUUUGCUAACCAAGUGC ...............................(((..((((((((...))))))))...))).(((.((((......)))).))).((((((((......)).....)))))) (-17.22 = -18.67 + 1.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:12 2011