| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,035,318 – 12,035,423 |
| Length | 105 |
| Max. P | 0.559009 |

| Location | 12,035,318 – 12,035,423 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 60.12 |
| Shannon entropy | 0.74633 |
| G+C content | 0.55597 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -7.85 |
| Energy contribution | -9.88 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
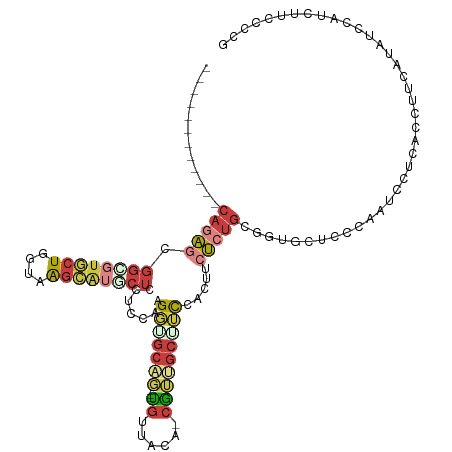
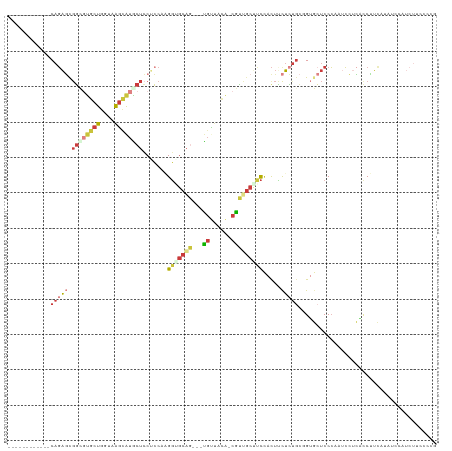
>dm3.chr2L 12035318 105 - 23011544 ------------CAGAGUGGCGUGCUGUUAAGCAUGCUCCGCCAAGGUGCAG---UGUUGCA-CGUUGCUUCCACUUCUCUGCGGUGCUCCCUAUCCUCACCUUCAUAUCCAUCUUCCCCG ------------(((((.((((((((....))))))))..((.((.((((..---....)))-).))))........))))).((((...........))))................... ( -28.90, z-score = -1.73, R) >droSim1.chr2L 11839308 105 - 22036055 ------------CAGAGCGGCGUGCUGUUAAGCAUACUCCUCCAAGGUGCAG---UGCUACA-CGUUGCUUCCACUUCUCUGCCGUGCUCCCUAUCCUCACCUUCAUAUCCAUCUUCCCCG ------------..((((((((((((....)))))..........((.((((---((.....-))))))..))........)))..))))............................... ( -20.90, z-score = -0.87, R) >droSec1.super_16 230447 105 - 1878335 ------------CAGAGCGGCGUGCUGUUAAGCAUACUCCUCCAAGGUGCAG---UGUUACA-CGUUGCAUCCACUUCUCUGCCGUGCUCCCCAUCCUCACCUUCAUAUCCAUCUUCCCCG ------------..((((((((((((....)))))..........(((((((---((.....-))))))))).........)))..))))............................... ( -23.70, z-score = -2.17, R) >droYak2.chr2L 8455447 105 - 22324452 ------------CAGAGCGGCGUGCUGGUCAACAUGCUCCUCCAUGGAGCAG---CGUUACA-CGUCGCUUCCACUUCUCUUCGGUGCUCCCAAUCCUCGUCUUCAUAUCCGUCUUCCCCU ------------..((((((((((......(((.((((((.....)))))).---.))).))-)))))))).((((.......)))).................................. ( -27.10, z-score = -1.82, R) >droEre2.scaffold_4929 13262627 108 + 26641161 ------------CAGAGCGGGGUGCUGGUAAGCAUGCUCCUCCAAGGAGCAGUGUUGUUACA-CGUUGCUUCUACUCCUCUGCGGUGCUCCCAGUCCUCACCUUCGUAUCCAUCUUCCCCU ------------.(((...((((((.((((((((((((((.....))))))((((....)))-)..)))))........(((.((....))))).....)))...)))))).)))...... ( -33.50, z-score = -1.34, R) >droAna3.scaffold_12916 6754102 90 + 16180835 ------------CAUAACGGUCCACUCGUCAGUAUGCUGCGCCAUGGCGC-----UGCAGGAUCACUGCCGCCAUUCCUUGG--GGACUCCAAGGGUCUGUCUUCUUCG------------ ------------....((((.((...((.(((....)))))..((((((.-----.((((.....))))))))))..(((((--.....))))))).))))........------------ ( -29.00, z-score = -0.76, R) >anoGam1.chr3R 37588 112 - 53272125 UAGGGAGGAGUACAGUCCAUUAUCCUUGAUGGAGAACCGCAGCACGUCACUCAUGUACUUGGGCAGAUCGGUAACCAUGAUGAAGAAACCCCAAUCAUGUCCGAUCUGUGCG--------- .......(((((((.(((((((....)))))))((...((.....))...)).)))))))(.(((((((((....((((((............)))))).))))))))).).--------- ( -36.20, z-score = -1.82, R) >consensus ____________CAGAGCGGCGUGCUGGUAAGCAUGCUCCUCCAAGGUGCAG___UGUUACA_CGUUGCUUCCACUUCUCUGCGGUGCUCCCAAUCCUCACCUUCAUAUCCAUCUUCCCCG ..............((((((((((((....)))))))).......(((((((.............)))))))..............))))............................... ( -7.85 = -9.88 + 2.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:18 2011