
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,649,603 – 19,649,735 |
| Length | 132 |
| Max. P | 0.774248 |

| Location | 19,649,603 – 19,649,703 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 58.61 |
| Shannon entropy | 0.77823 |
| G+C content | 0.32396 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -6.34 |
| Energy contribution | -7.07 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
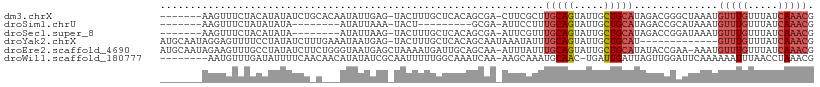
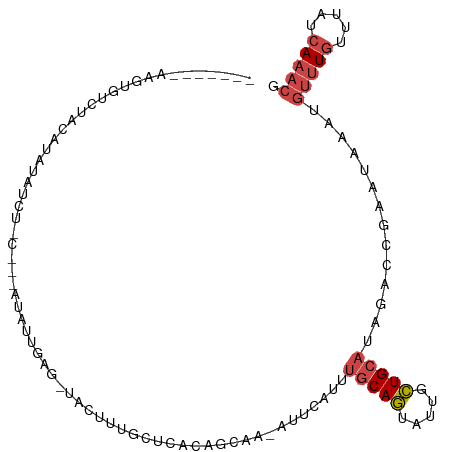
>dm3.chrX 19649603 100 + 22422827 -------AAGUUUCUACAUAUAUCUGCACAAUAUUGAG-UACUUUGCUCACAGCGA-CUUCGCUUGCAGUAUUGCUGCAUAGACGGGCUAAAUGUUUGUUUAUCAAACG -------..((((............(((((((((((((-(.....))))..((((.-...))))...))))))).)))((((((((((.....)))))))))).)))). ( -25.70, z-score = -1.86, R) >droSim1.chrU 11162349 83 + 15797150 -------AAGUUUCUAUAUAUA--------AUAUUAAA-UACU---------GCGA-AUUCCUUUGCAGUAUUGCUGCAUAGACCGCAUAAAUGUUUGUUUAUCAAACG -------...............--------......((-((((---------((((-(....)))))))))))..(((.......))).....(((((.....))))). ( -16.90, z-score = -2.30, R) >droSec1.super_8 1924448 92 + 3762037 -------AAGUUUCUACAUAUA--------AUAUUAAG-UACUUUGCUCACAGCGA-AUUCGUUUGCAGUAUUGCUGCAUAGACCGGAUAAAUGUUUGUUUAUCAAACG -------..(((((((......--------........-...(((((.....))))-)......(((((.....)))))))))...(((((((....))))))).))). ( -16.70, z-score = -0.53, R) >droYak2.chrX 10071295 95 - 21770863 AUGCAAUAGGAGUUUUCCUAUAUCUUUGAAAUAAUGAG-UACUUUGCUCACAGCAAUAAAUAUUUGCAGUAUUGCUGCAU-------------GUUUGUUUAUCAAACG .(((.(((((......))))).............((((-(.....)))))..))).........(((((.....))))).-------------(((((.....))))). ( -23.00, z-score = -2.22, R) >droEre2.scaffold_4690 9871382 107 + 18748788 AUGCAAUAGAAGUUUGCCUAUAUCUUCUGGGUAAUGAGCUAAAAUGAUUGCAGCAA-AUUUAUUUGCAGUAUUGCUGCAUAUACCGAA-AAAUGUUUGUUUAUCAAACG .((((((...((((((((((.......)))))...)))))......))))))....-...(((.(((((.....))))).))).....-....(((((.....))))). ( -22.70, z-score = -0.69, R) >droWil1.scaffold_180777 3692297 99 - 4753960 --------AAUGUUUGAUAUUUUCAACAACAUAUAUCGCAAUUUUUGGCAAAUCAA-AAGCAAAUGCAAC-UGAUUGAUUAGUUGGAUUCAAAAAAUUUAACCUAAACG --------....(((((....((((((..........((........)).(((((.-..((....))...-))))).....)))))).)))))................ ( -13.90, z-score = -0.30, R) >consensus _______AAGUGUCUACAUAUAUCU_C___AUAUUGAG_UACUUUGCUCACAGCAA_AUUCAUUUGCAGUAUUGCUGCAUAGACCGAAUAAAUGUUUGUUUAUCAAACG ................................................................(((((.....)))))..............(((((.....))))). ( -6.34 = -7.07 + 0.72)
| Location | 19,649,634 – 19,649,735 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 63.28 |
| Shannon entropy | 0.66600 |
| G+C content | 0.36766 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -9.27 |
| Energy contribution | -8.80 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
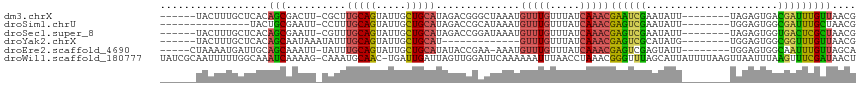
>dm3.chrX 19649634 101 + 22422827 ------UACUUUGCUCACAGCGACUU-CGCUUGCAGUAUUGCUGCAUAGACGGGCUAAAUGUUUGUUUAUCAAACGAAUCGAAUAUU--------UAGAGUGACGAUUUGUUAACG ------............((((....-))))(((((.....)))))(((((((((.....)))))))))...(((((((((......--------........))))))))).... ( -25.34, z-score = -1.29, R) >droSim1.chrU 11162372 92 + 15797150 ---------------UACUGCGAAUU-CCUUUGCAGUAUUGCUGCAUAGACCGCAUAAAUGUUUGUUUAUCAAACGAGUCGAAUAUU--------UGGAGUGGCGAUUUGCUAACG ---------------....(((((((-.((.(((((.....))))).)).((((.((((((((((....((....))..))))))))--------))..)))).)))))))..... ( -26.80, z-score = -2.57, R) >droSec1.super_8 1924471 101 + 3762037 ------UACUUUGCUCACAGCGAAUU-CGUUUGCAGUAUUGCUGCAUAGACCGGAUAAAUGUUUGUUUAUCAAACGAGUCGAAUAUU--------UAGAGUGGUGACUCGCUAACG ------............(((((.((-(((((((((.....))))........(((((((....)))))))))))))))).......--------..((((....))))))).... ( -25.10, z-score = -0.95, R) >droYak2.chrX 10071333 89 - 21770863 ------UACUUUGCUCACAGCAAUAAAUAUUUGCAGUAUUGCUGCAU-------------GUUUGUUUAUCAAACGAGUCGCAUAUG--------UGGAGUGGCGGUUUGUUAACG ------....((((.....))))((((((..(((((.....))))).-------------...))))))...(((((..(((.(((.--------....))))))..))))).... ( -22.10, z-score = -0.45, R) >droEre2.scaffold_4690 9871420 101 + 18748788 -----CUAAAAUGAUUGCAGCAAAUU-UAUUUGCAGUAUUGCUGCAUAUACCGAA-AAAUGUUUGUUUAUCAAACGAGUCGAGUAUU--------UGGAGUGGCAAUUUGUUAGCA -----((((...((((((.(((((..-..)))))......(((.((.(((((((.-...((((((.....))))))..))).)))).--------)).))).))))))..)))).. ( -28.20, z-score = -2.67, R) >droWil1.scaffold_180777 3692322 114 - 4753960 UAUCGCAAUUUUUGGCAAAUCAAAAG-CAAAUGCAAC-UGAUUGAUUAGUUGGAUUCAAAAAAUUUAACCUAAACGGGUUUAGCAUUAUUUUAAGUUAAUUUAAGUUUCGAUAACU (((((.((((((((((..........-..(((.((((-(((....))))))).)))..........(((((....)))))..............)))))...))))).)))))... ( -18.40, z-score = -0.59, R) >consensus ______UACUUUGCUCACAGCAAAUU_CAUUUGCAGUAUUGCUGCAUAGACCGAAUAAAUGUUUGUUUAUCAAACGAGUCGAAUAUU________UGGAGUGGCGAUUUGUUAACG ..................(((..........(((((.....)))))..............(((((.....)))))((((((......................))))))))).... ( -9.27 = -8.80 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:02:01 2011