| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,026,256 – 12,026,362 |
| Length | 106 |
| Max. P | 0.977204 |

| Location | 12,026,256 – 12,026,362 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.41 |
| Shannon entropy | 0.50827 |
| G+C content | 0.50735 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
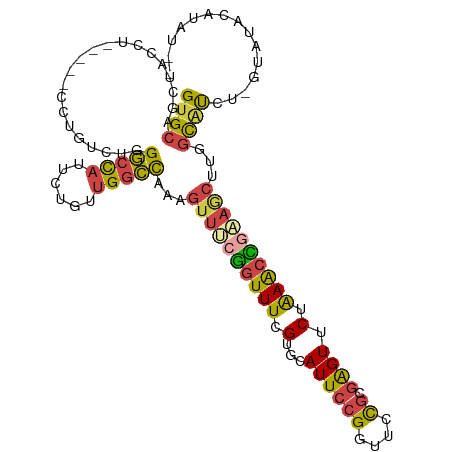
>dm3.chr2L 12026256 106 + 23011544 GUGCAGCUACCUU----UCUGUCUGGGCCAUUCUGUUGGCCAAAGUUUCGGUUUCGUGCAUUCCGGUUCUGCGAGUUCUAAACCGAAGCUUGGCAUCU-GUAUACAUAUAC ((((((...((..----........(((((......))))).((((((((((((.(...((((((....)).)))).).))))))))))))))...))-))))........ ( -33.30, z-score = -2.64, R) >droAna3.scaffold_12916 6741625 98 - 16180835 GUGCAGGAGGUG--------GAAAGGAGUUUCCAUCUGGUUGGA--CCAACUUUGGCAGAUUCAGGUUUCGCUAGUUCUAAAUUGGAGCAGGGUGCAUUGUAUUCCAC--- (((((...((((--------(((..(((((((((...(((((..--.))))).))).))))))...))))))).(((((.....)))))....)))))..........--- ( -27.50, z-score = -0.51, R) >droEre2.scaffold_4929 13253565 102 - 26641161 GUGCAGCUACCUU----CCAGUCUGGGCCAUUCUUUUGGCCAAUGUUUCGGUUUCGGGCAUUCCGGUUCCGCGAGUUCUGAACUGAAACUGGCUGCAC-ACAUGCAU---- (((((((((....----........(((((......)))))...(((((((((.((((.((((((....)).))))))))))))))))))))))))))-........---- ( -44.00, z-score = -4.60, R) >droYak2.chr2L 8445839 101 + 22324452 GUGCAGCUACCU-----CCUGUCUGGGCCAUUCUGUUGGCCAAAGUUUCGGUUUCGUGCAUUCCGGUUCCGCGGGUUCUAAGCCCAAGCU-GGC-UGC-ACAUACAUAU-- ((((((((....-----........(((((......)))))..((((((((..(((.......)))..))).((((.....)))))))))-)))-)))-))........-- ( -37.60, z-score = -2.62, R) >droSec1.super_16 221649 106 + 1878335 GUGCAGCUACCU----GCCUGUCUGGGCCAUUCUGUUGGCCAAAGUUUCGGUUUCGUGCAUUCCGGUUCUGCGAGUUCUAAACCGCAGCUUGGCAUCU-GUAUACAUAUAC ((((((.....(----(((......(((((......))))).(((((.((((((.(...((((((....)).)))).).)))))).))))))))).))-))))........ ( -31.70, z-score = -1.11, R) >droSim1.chr2L 11830366 110 + 22036055 GUGCAGCUACCUUCCUGUCUGUCUGGGCCAUUCUGUUGGCCAAAGUUUCGGUUUCGUGCAUUCCGGUUCUGCGAGUUCUAAACCGCAGCUUGGCAUCU-GUAUACAUAUAC ((((((...((..............(((((......))))).(((((.((((((.(...((((((....)).)))).).)))))).)))))))...))-))))........ ( -28.90, z-score = -0.62, R) >consensus GUGCAGCUACCU_____CCUGUCUGGGCCAUUCUGUUGGCCAAAGUUUCGGUUUCGUGCAUUCCGGUUCCGCGAGUUCUAAACCGAAGCUUGGCAUCU_GUAUACAUAU__ ((((.....................(((((......)))))...((((((((((.(...((((((....)).)))).).))))))))))...))))............... (-18.70 = -18.57 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:17 2011