
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,646,757 – 19,646,863 |
| Length | 106 |
| Max. P | 0.967583 |

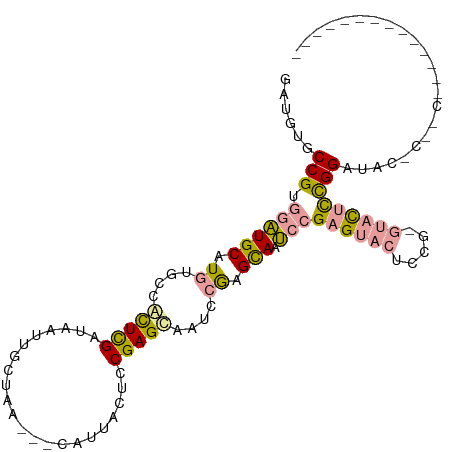
| Location | 19,646,757 – 19,646,863 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 61.18 |
| Shannon entropy | 0.53282 |
| G+C content | 0.51437 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -10.62 |
| Energy contribution | -11.08 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
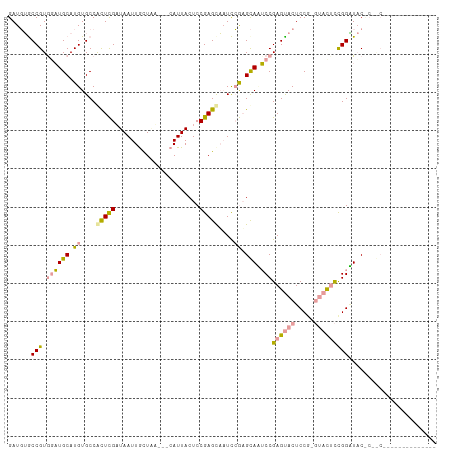
>dm3.chrX 19646757 106 + 22422827 GAUCUGCCGUGGCUGCAUGUGCCACUCGUUAAUUGCUAA---CAUUACUCCGAGUACUCGGAGUAUGCCGAGCACUCCG---AUGUUGGAUACCCAACCGGUGGCCAUCACU (((..((((((((.......)))))........((((..---...((((((((....)))))))).....))))..(((---..(((((....)))))))).))).)))... ( -39.60, z-score = -3.28, R) >droYak2.chrX 10068609 98 - 21770863 GAUGUGCCGUGUAUGCAUGUGCCAUUUGAUAAUUACUCC---CAUUACUCCGAGCAAUCCAUGCACUCCGAGUACUCCAUGUACUCCGGGUACUCCGCACU----------- ...((((.(.((((((((((((..((.((((((......---.)))).)).)))))...)))))...((((((((.....))))).))))))).).)))).----------- ( -25.90, z-score = -2.00, R) >droEre2.scaffold_4690 9868829 93 + 18748788 GAUGAACCGUAGAUGCAUGUGCCGCUCGAUAAUUGUUGUUGGCAUUACUCCGAGCUAUUCGAGCAAUCCGAGUACUCCGAGUACUCCGGUUCC------------------- ...((((((.((......((((((..((((....)))).))))))(((((.(((.((((((.......))))))))).))))))).)))))).------------------- ( -32.70, z-score = -3.56, R) >consensus GAUGUGCCGUGGAUGCAUGUGCCACUCGAUAAUUGCUAA___CAUUACUCCGAGCAAUCCGAGCAAUCCGAGUACUCCG_GUACUCCGGAUAC_C__C______________ ......(((.((((((.((....(((((......................)))))....)).))).)))((((((.....)))))))))....................... (-10.62 = -11.08 + 0.46)

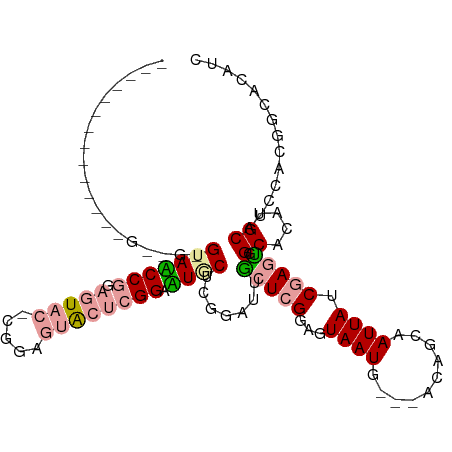
| Location | 19,646,757 – 19,646,863 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 61.18 |
| Shannon entropy | 0.53282 |
| G+C content | 0.51437 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -10.34 |
| Energy contribution | -11.57 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
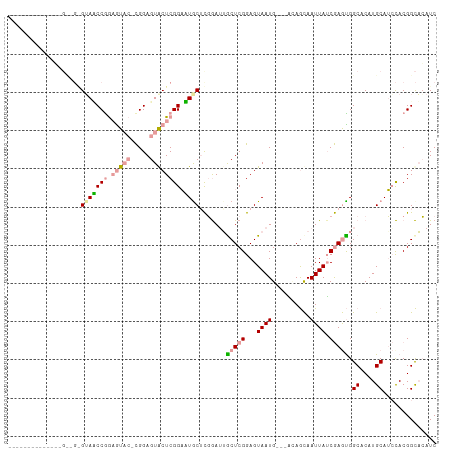
>dm3.chrX 19646757 106 - 22422827 AGUGAUGGCCACCGGUUGGGUAUCCAACAU---CGGAGUGCUCGGCAUACUCCGAGUACUCGGAGUAAUG---UUAGCAAUUAACGAGUGGCACAUGCAGCCACGGCAGAUC .((.(((((((((((((((....))))).(---(.((((((((((......)))))))))).))......---...........)).))))).))))).((....))..... ( -42.70, z-score = -3.34, R) >droYak2.chrX 10068609 98 + 21770863 -----------AGUGCGGAGUACCCGGAGUACAUGGAGUACUCGGAGUGCAUGGAUUGCUCGGAGUAAUG---GGAGUAAUUAUCAAAUGGCACAUGCAUACACGGCACAUC -----------.((((.(.((..(((.(((((.....)))))))).(((((((((((((((..(....).---.)))))))).((....))..))))))))).).))))... ( -33.70, z-score = -2.77, R) >droEre2.scaffold_4690 9868829 93 - 18748788 -------------------GGAACCGGAGUACUCGGAGUACUCGGAUUGCUCGAAUAGCUCGGAGUAAUGCCAACAACAAUUAUCGAGCGGCACAUGCAUCUACGGUUCAUC -------------------.((((((((((((.....))))))((((.((.......(((((..(((((..........)))))))))).......)))))).))))))... ( -31.44, z-score = -3.06, R) >consensus ______________G__G_GUAACCGGAGUAC_CGGAGUACUCGGAAUGCUCGGAUUGCUCGGAGUAAUG___ACAGCAAUUAUCGAGUGGCACAUGCAUCCACGGCACAUC ...................(((((((.(((((.....)))))))).)))).......(((((...((((..........)))).))))).((....)).............. (-10.34 = -11.57 + 1.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:57 2011