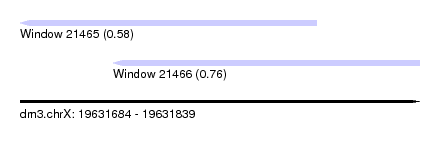
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,631,684 – 19,631,839 |
| Length | 155 |
| Max. P | 0.758399 |

| Location | 19,631,684 – 19,631,799 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 57.10 |
| Shannon entropy | 0.69986 |
| G+C content | 0.54877 |
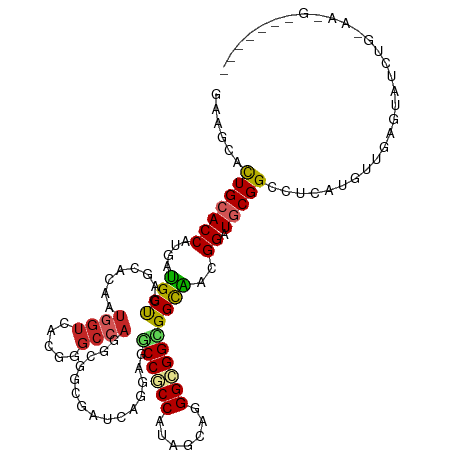
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -16.50 |
| Energy contribution | -15.88 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
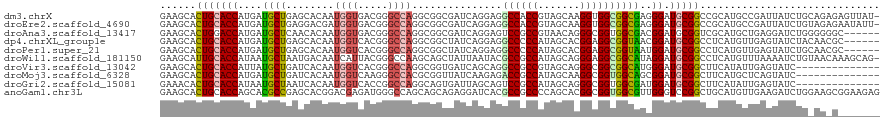
>dm3.chrX 19631684 115 - 22422827 CAGGCGGCGAUCAGGAGGCCACCGUAGCAAGGUGGCGGCGAGGGAUGCGGCCGCAUGCCGAUUAUCU-GCAGAGAGUUAUAUAAUUCCAUCAAUGCUUUCUCCACCCUCCGUAUCA-- ........(((..((((((((((.......))))))((.((((.((((....)))).))........-(((..(((((....)))))......)))..)).))...))))..))).-- ( -36.70, z-score = 0.31, R) >droEre2.scaffold_4690 9853856 107 - 18748788 CAGGCGGCGAUCAGGAGGCCACCGUAGCAAGGUGGCGGCGAGGGAUGCGGCCGCAUGCCGAUUAUCU-GUAGAGAAUAUUUUAUUUCCAUCAAAAUUCAAUCCUGCCA---------- ..(((((.(((...(((((((((.......))))))((.((((.((((....)))).)).....(((-....)))........)).)).......))).)))))))).---------- ( -35.00, z-score = -0.62, R) >droWil1.scaffold_181150 1759772 116 - 4952429 CAAGCAGCUAUUAAUACGCCGCCAUAGCAGGGAGGCGGCAUAGGAUGCGGCCUCAUGUUUAAAAUCU-GUAACAAAGCAGGCAGAGAU-UAGGUACACAGUUCACCCACUCCGUCUUA ......(((.....((((.......((((..(((((.((((...)))).))))).)))).......)-)))....)))((((.(((..-..(((.........)))..))).)))).. ( -27.94, z-score = 0.37, R) >anoGam1.chr3L 30803571 104 - 41284009 CAGCAGCAGAGGAUCACGCCGCCCCAGCACGGCGGUGGCGUUGGGUCCGGCUGCAUGUUGAAGAUCUGGAAGCGGAAGAGACAUUU---UAGACGCGGAAACCUCUG----------- ..(((((...(((((((((((((((.....)).))))))))..))))).)))))(((((.....((((....))))...)))))..---((((.(.(....))))))----------- ( -43.90, z-score = -2.56, R) >consensus CAGGCGGCGAUCAGGACGCCACCGUAGCAAGGUGGCGGCGAGGGAUGCGGCCGCAUGCCGAAUAUCU_GUAGAGAAUAAGACAUUUCC_UAAAUACUCAAUCCACCCA__________ ..((((...........((((((.......))))))(((..........)))...))))........................................................... (-16.50 = -15.88 + -0.62)


| Location | 19,631,720 – 19,631,839 |
|---|---|
| Length | 119 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.88 |
| Shannon entropy | 0.56236 |
| G+C content | 0.58782 |
| Mean single sequence MFE | -42.42 |
| Consensus MFE | -21.77 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19631720 119 - 22422827 GAAGCACUGCACCAUGAUGCUGAGCACAAUGGUGACGGGCCAGGCGGCGAUCAGGAGGCCACCGUAGCAAGGUGGCGGCGAGGGAUGCGGCCGCAUGCCGAUUAUCUGCAGAGAGUUAU- ...((.(((((((((..(((...)))..)))))).)))))...((((.((((..(..((((((.......))))))..)..((.((((....)))).))))))..))))..........- ( -45.60, z-score = -0.80, R) >droEre2.scaffold_4690 9853884 119 - 18748788 GAAGCACUGCACCAUGAUGCUGAGGACGAUGGUGACGGGCCAGGCGGCGAUCAGGAGGCCACCGUAGCAAGGUGGCGGCGAGGGAUGCGGCCGCAUGCCGAUUAUCUGUAGAGAAUAUU- ......(((((((.((((((((.((....((....))..))...)))).))))))..((((((.......)))))).....((.((((....)))).)).......)))))........- ( -41.30, z-score = -0.22, R) >droAna3.scaffold_13417 4752221 114 + 6960332 GAAGCACUGGACCAUGAUGCUCAACACAAUGGUGACGGGCCAGGCGGCGAUCAGGAGUCCGCCGUAACAGGGCGGUGGCGACGGAUGCGGUCGCAUGCUGAGGAUCUGGGGGGC------ ...((.((...(((.(((.((((.(((..((....)).(((..((((((..(....)..)))))).....))).)))(((((.......)))))....)))).)))))).))))------ ( -42.20, z-score = -0.20, R) >dp4.chrXL_group1e 6521827 114 - 12523060 GAAGCACUGCACCAUGAUGCUGAGCACAAUGGUCACGGGCCAGGCGGCUAUCAGGAGGCCCCCAUAGCACGGAGGCGGUAACGGAUGCGGCCUCAUGUUGAGUAUCUACAACGC------ ...............((((((.((((...((((.....))))..(((((((..((....))..))))).))(((((.(((.....))).))))).)))).))))))........------ ( -36.90, z-score = 0.58, R) >droPer1.super_21 1530305 114 - 1645244 GAAGCACUGCACCAUGAUGCUGAGCACAAUGGUCACGGGCCAGGCGGCUAUCAGGAGGCCCCCAUAGCACGGAGGCGGUAAUGGAUGCGGCCUCAUGUUGAGUAUCUGCAACGC------ ...((..((((....((((((.((((...((((.....))))..(((((((..((....))..))))).))(((((.(((.....))).))))).)))).))))))))))..))------ ( -40.60, z-score = 0.11, R) >droWil1.scaffold_181150 1759809 119 - 4952429 GAAGCAUUGCACCAUAAUGCUAAUGACAAUCAUUACGGGCCAAGCAGCUAUUAAUACGCCGCCAUAGCAGGGAGGCGGCAUAGGAUGCGGCCUCAUGUUUAAAAUCUGUAACAAAGCAG- ..(((((((.....)))))))((((.....))))..(((((..(((.((((......((((((..........))))))))))..))))))))............((((......))))- ( -32.00, z-score = -0.41, R) >droVir3.scaffold_13042 4325554 106 + 5191987 GAAGCACUGCACCAUUAUGCUGAUCACAAUGGUCACGGGCCAGGCGGUGAUCAGCAGGCCGCCGUAGCAGGGCGGCGGCAUGGGAUGCGGCUUCAUAUUGAGUAUC-------------- (((((...(((((....((((((((((..((((.....))))....)))))))))).((((((((......))))))))...)).))).)))))............-------------- ( -53.80, z-score = -5.10, R) >droMoj3.scaffold_6328 1594521 106 + 4453435 GAAGCACUGCACCAUGAUGCUGAUCACAAUGGUCAAGGGCCACGCGGUUAUCAAGAGACCGCCAUAGCAAGGCGGUGGCAGCGGAUGCGGCUUCAUGCUCAGUAUC-------------- .....((((...(((((.(((((((....((((.....))))(((((((.......))))(((((.((...)).))))).)))))).)))).)))))..))))...-------------- ( -38.70, z-score = -1.37, R) >droGri2.scaffold_15081 3026130 106 + 4274704 GAAACACUGCACCAUAAUGCUAAUCACAAUGGUCACCGGCCAGGCAGUGAUUAGCAGUCCGCCAUAGCAGGGCGGUGGCGAUGGAUGCGGCUUCAUAUUGAGUAUC-------------- (((...(((((((((..((((((((((..((((.....))))....))))))))))...((((((.((...)).)))))))))).))))).)))............-------------- ( -41.70, z-score = -2.97, R) >anoGam1.chr3L 30803595 120 - 41284009 GAAGCACUGCACCAGCACGCCGAGCACGGACGAGAUGGGCCAGCAGCAGAGGAUCACGCCGCCCCAGCACGGCGGUGGCGUUGGGUCCGGCUGCAUGUUGAAGAUCUGGAAGCGGAAGAG ......((((.(((.(.(((((....))).)).).)))..(((((((((.(((((((((((((((.....)).))))))))..)))))..)))).)))))...........))))..... ( -51.40, z-score = -1.66, R) >consensus GAAGCACUGCACCAUGAUGCUGAGCACAAUGGUCACGGGCCAGGCGGCGAUCAGGAGGCCGCCAUAGCAGGGCGGCGGCAACGGAUGCGGCCUCAUGUUGAGUAUCUG_AA_G_______ ......(((((((....((((........((((.....))))...............((((((.......))))))))))..)).))))).............................. (-21.77 = -21.90 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:54 2011