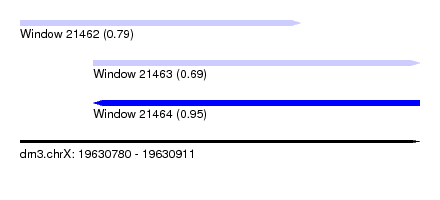
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,630,780 – 19,630,911 |
| Length | 131 |
| Max. P | 0.949528 |

| Location | 19,630,780 – 19,630,872 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 53.48 |
| Shannon entropy | 0.72293 |
| G+C content | 0.51533 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -9.76 |
| Energy contribution | -13.08 |
| Covariance contribution | 3.32 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
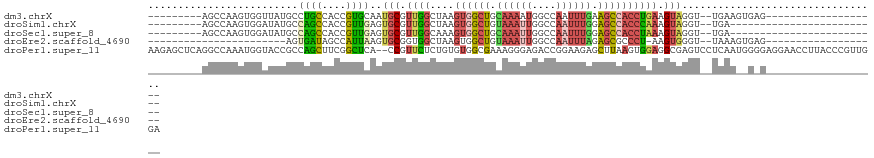
>dm3.chrX 19630780 92 + 22422827 ---------AGCCAAGUGGUUAUGCCUGCCACCGUGCAAUGCGUUGGCUAAGUGGCUGCAAAAUGGCCAAUUUGAAGCCACCUGAAGUAGGU--UGAAGUGAG------------------- ---------((((....))))..((((((((.(((.....))).))))...((((((.((((........)))).)))))).......))))--.........------------------- ( -26.80, z-score = -0.00, R) >droSim1.chrX 15147902 86 + 17042790 ---------AGCCAAGUGGAUAUGCCAGCCACCGUUGAGUGCGUUGGCUAAGUGGCUGUAAAUUGGCCAAUUUGGAGCCACCCAAAGUAGGU--UGA------------------------- ---------((((...(((....(((((((((......))).))))))...((((((.((((((....)))))).))))))))).....)))--)..------------------------- ( -33.30, z-score = -2.76, R) >droSec1.super_8 1905754 86 + 3762037 ---------AGCCAAGUGGAUAUGCCAGCCACCGUUGAGUGCGUUGGCAAAGUGGCUGCAAAUUGGCCAAUUUGGAGCCACCUAAAGUAGGU--UGA------------------------- ---------((((.........((((((((((......))).)))))))..((((((.((((((....)))))).))))))........)))--)..------------------------- ( -34.50, z-score = -2.85, R) >droEre2.scaffold_4690 9853003 77 + 18748788 -----------------------AGUGAUAGCCAUUAAGUGCGGUGGCUAAGUGGCUGUAAAUUGGCCAAUUUAGAGCGCCCU-AAGUGGGU--UAAAGUGAG------------------- -----------------------....(((((((((.(((......))).)))))))))...((((((.((((((......))-)))).)))--)))......------------------- ( -23.80, z-score = -1.83, R) >droPer1.super_11 133114 120 + 2846995 AAGAGCUCAGGCCAAAUGGUACCGCCAGCUUCGGCUCA--CCGUUCUCUGUGUGGCGAAAGGGAGACCGGAAGAGCUUAAGUGGAGGCGAGUCCUCAAUGGGGAGGAACCUUACCCGUUGGA .((..(((..(((....)))...(((.((((.(((((.--(((.(((((.(........).))))).)))..))))).))))...))))))..))((((((((((....))).))))))).. ( -48.40, z-score = -1.46, R) >consensus _________AGCCAAGUGGAUAUGCCAGCCACCGUUCAGUGCGUUGGCUAAGUGGCUGCAAAUUGGCCAAUUUGGAGCCACCUAAAGUAGGU__UGAA__G_G___________________ .......................(((((((((......))).))))))...((((((.((((((....)))))).))))))......................................... ( -9.76 = -13.08 + 3.32)

| Location | 19,630,804 – 19,630,911 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 59.26 |
| Shannon entropy | 0.70053 |
| G+C content | 0.51302 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -13.34 |
| Energy contribution | -12.98 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19630804 107 + 22422827 GUGCAAUGCGUUGGCUAAGUGGCUGCAAAAUGGCCAAUUUGAAGCC--ACCUGAAGUAGGUUGAAGUGAGUUACUGCUCACUGCUCUGGCCAGUGACCAGAAGUGAGCA-- .........(.(((((...(((((.......)))))......))))--).)...(((((.((.....)).)))))(((((((..(((((.......)))))))))))).-- ( -33.60, z-score = -0.18, R) >droSim1.chrX 15147926 83 + 17042790 GUUGAGUGCGUUGGCUAAGUGGCUGUAAAUUGGCCAAUUUGGAGCC--ACCCAAAGUAGGUUGA------------------------GCCAGUGACCAGAAGUGAGCA-- .(((.((.((((((((..((((((.((((((....)))))).))))--))((......))...)------------------------)))))))))))).........-- ( -26.30, z-score = -1.62, R) >droSec1.super_8 1905778 83 + 3762037 GUUGAGUGCGUUGGCAAAGUGGCUGCAAAUUGGCCAAUUUGGAGCC--ACCUAAAGUAGGUUGA------------------------GCCAGUGACCAGAAGUGAGCA-- .(((.((.(((((((.....((((.((((((....)))))).))))--(((((...)))))...------------------------)))))))))))).........-- ( -25.90, z-score = -1.12, R) >droEre2.scaffold_4690 9853014 98 + 18748788 -UUAAGUGCGGUGGCUAAGUGGCUGUAAAUUGGCCAAUUUAGAGC---GCCCUAAGUGGGUUAAAGUGAGU-------CACUACUCUGGCCAGUGACCAGAAGUGAGCA-- -.....((((((((((.....(((.((((((....)))))).)))---((((.....)))).......)))-------))))(((((((.......)))).)))..)))-- ( -31.20, z-score = -1.01, R) >droPer1.super_11 133154 111 + 2846995 GUUCUCUGUGUGGCGAAAGGGAGACCGGAAGAGCUUAAGUGGAGGCGAGUCCUCAAUGGGGAGGAACCUUACCCGUUGGAUCGCUGCUCUCCCGUAUCACAUGAUCGCACU ((..((((((..(((...((....))(((.((((.........(((((....(((((((((((....))).)))))))).)))))))))))))))..)))).))..))... ( -40.30, z-score = -0.87, R) >consensus GUUCAGUGCGUUGGCUAAGUGGCUGCAAAUUGGCCAAUUUGGAGCC__ACCCAAAGUAGGUUGAA______________A___CU___GCCAGUGACCAGAAGUGAGCA__ ......(((..((((.....((((.((((((....)))))).))))..((((.....))))...........................)))).(.((.....)).)))).. (-13.34 = -12.98 + -0.36)

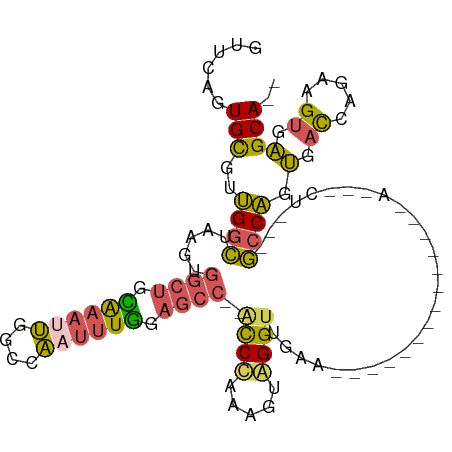
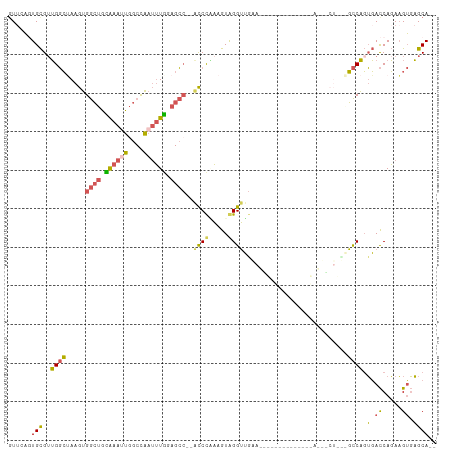
| Location | 19,630,804 – 19,630,911 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 59.26 |
| Shannon entropy | 0.70053 |
| G+C content | 0.51302 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -11.16 |
| Energy contribution | -10.64 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
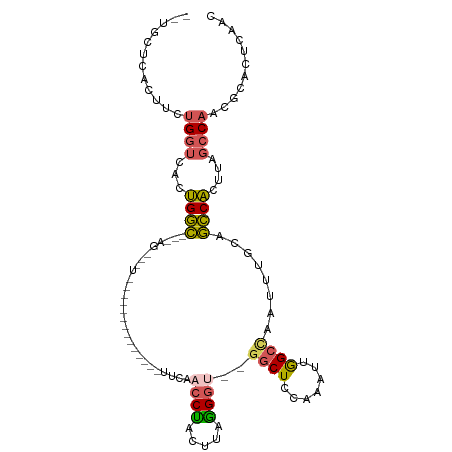
>dm3.chrX 19630804 107 - 22422827 --UGCUCACUUCUGGUCACUGGCCAGAGCAGUGAGCAGUAACUCACUUCAACCUACUUCAGGU--GGCUUCAAAUUGGCCAUUUUGCAGCCACUUAGCCAACGCAUUGCAC --((((((((((((((.....))))))..))))))))((((..................((((--((((.((((........)))).)))))))).((....)).)))).. ( -38.60, z-score = -3.29, R) >droSim1.chrX 15147926 83 - 17042790 --UGCUCACUUCUGGUCACUGGC------------------------UCAACCUACUUUGGGU--GGCUCCAAAUUGGCCAAUUUACAGCCACUUAGCCAACGCACUCAAC --((((((....)))....((((------------------------(...((......))((--((((..(((((....)))))..))))))..)))))..)))...... ( -19.90, z-score = -1.12, R) >droSec1.super_8 1905778 83 - 3762037 --UGCUCACUUCUGGUCACUGGC------------------------UCAACCUACUUUAGGU--GGCUCCAAAUUGGCCAAUUUGCAGCCACUUUGCCAACGCACUCAAC --((((((....)))....((((------------------------............((((--((((.((((((....)))))).)))))))).))))..)))...... ( -21.12, z-score = -1.37, R) >droEre2.scaffold_4690 9853014 98 - 18748788 --UGCUCACUUCUGGUCACUGGCCAGAGUAGUG-------ACUCACUUUAACCCACUUAGGGC---GCUCUAAAUUGGCCAAUUUACAGCCACUUAGCCACCGCACUUAA- --((((((((((((((.....))))))..))))-------)..........(((.....))).---(((.((((((....)))))).)))............))).....- ( -24.60, z-score = -1.02, R) >droPer1.super_11 133154 111 - 2846995 AGUGCGAUCAUGUGAUACGGGAGAGCAGCGAUCCAACGGGUAAGGUUCCUCCCCAUUGAGGACUCGCCUCCACUUAAGCUCUUCCGGUCUCCCUUUCGCCACACAGAGAAC .((((((....(.(((.(((((((((..((......))((..(((((((((......)))))...))))))......)))))))))))).)....))).)))......... ( -36.50, z-score = -1.32, R) >consensus __UGCUCACUUCUGGUCACUGGC___AG___U______________UUCAACCUACUUAGGGU__GGCUCCAAAUUGGCCAAUUUGCAGCCACUUAGCCAACGCACUCAAC ............((((...((((............................(((.....)))...((((.......))))........))))....))))........... (-11.16 = -10.64 + -0.52)

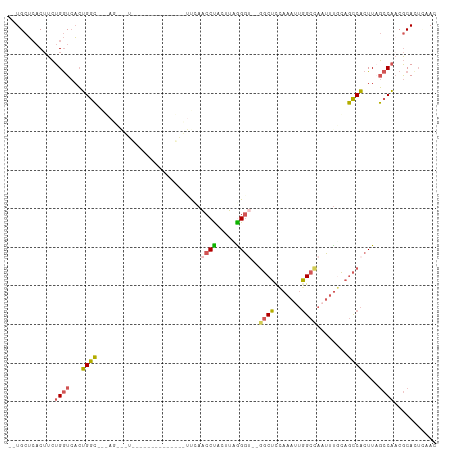
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:53 2011