| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,594,993 – 19,595,085 |
| Length | 92 |
| Max. P | 0.519089 |

| Location | 19,594,993 – 19,595,085 |
|---|---|
| Length | 92 |
| Sequences | 13 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 70.40 |
| Shannon entropy | 0.65899 |
| G+C content | 0.62472 |
| Mean single sequence MFE | -36.81 |
| Consensus MFE | -16.83 |
| Energy contribution | -16.68 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.95 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19594993 92 + 22422827 CGCUGCUGGGCAAGUGGCAGCGGAAACUCGUAGGCCAGCGGAUCGCUGGCCAGCUGGGCGGCAUGCUCAAUGGCCCGUCCUCC----GUUCCACAA .(((....)))..((((.((((((.....((.((((((((...)))))))).)).((((((...(((....))))))))))))----))))))).. ( -45.40, z-score = -1.89, R) >droSim1.chrX 15089479 92 + 17042790 CGCUGCUGGGCGAGUGGCAGCGGAAACUCGUAGGCCAGCGGAUCGCUGGCCAGCUGGGCGGCAUGCUCAAUGGCCCGUCCUCC----GUUCCACAA ((((....)))).((((.((((((.....((.((((((((...)))))))).)).((((((...(((....))))))))))))----))))))).. ( -46.90, z-score = -2.03, R) >droSec1.super_8 1871697 92 + 3762037 CGCUGCUGGGCGAGUGGCAGCGGAAACUCGUAGGCCAGCGGAUCGCUGGCCAGCUGGGCGGCAUGCUCAAUGGCCCGUCCUCC----GUUCCACAA ((((....)))).((((.((((((.....((.((((((((...)))))))).)).((((((...(((....))))))))))))----))))))).. ( -46.90, z-score = -2.03, R) >droYak2.chrX 10022527 92 - 21770863 CGCUGCUGAGCGAGUGGCAGCGGAAACUCGUACGCCAGCGGAUCGCUGGCCAGCUGGGCGGCGUGCUCAAUCGCCCGUCCUCC----AUUCCACAG .((((((........))))))((((........(((((((...)))))))..(..((((((.(((......)))))))))..)----.)))).... ( -40.00, z-score = -0.97, R) >droEre2.scaffold_4690 9819338 92 + 18748788 CGCUGCUGGGCGAGUGGCAGCGGAAACUCGUAGGCUAACGGAUUGCUGGCCAGCUGGGCGGCGUGUUCAAUGGCUCGUCCUCC----GUUCCACAG ((((....)))).((((.((((((....((..(((((..(((((((((.((....)).))))).))))..)))))))...)))----))))))).. ( -36.40, z-score = -0.13, R) >droAna3.scaffold_13417 4710945 92 - 6960332 CGUUGCUGGGACAGCGGCAGGGGGAAAUCGUAGGCCGAGGGAUCUGUGGCCAGGCGAGCAGCAUGUUCGAUGGCCCUGCCACC----GUUCCACAG ...((.((((((.(.(((((((..........(((((((....)).)))))...(((((.....)))))....))))))).).----)))))))). ( -39.70, z-score = -1.21, R) >dp4.chrXL_group1e 956040 92 + 12523060 CGCUGCUGUGCAAGCGGCAGGGGGAAGUCGUAGGCGUCUGGACAGACAGCCGAUUGAGCGGCAUGCUCGAUGGCCUGCCCACC----AUUCCAUGC (.(((((((....))))))).)((((...((.(((((((....)))).(((.(((((((.....))))))))))..))).)).----.)))).... ( -40.60, z-score = -1.29, R) >droPer1.super_30 715032 92 - 958394 CGCUGCUGGGCAAGCGGUAGGGGGAAGUCGUAGGCGUCUGGACAGACAGCCGAUUGAGCGGCAUGCUCGAUGGCCUGCCCACC----AUUCCAUGC (.((((((......)))))).)((((...((.(((((((....)))).(((.(((((((.....))))))))))..))).)).----.)))).... ( -36.90, z-score = -0.37, R) >droWil1.scaffold_181150 1717017 92 + 4952429 CUUUGCUGAGCCAACGGCAGUGGGAAAUCAUAGGCUAGUGGAUCUUUGGCAGACUUUGCGGCAUGCUCAAUGGCACGACCACC----GUUCCAUAG ......(((((...(.(((((((....))))..(((((.......)))))......))).)...)))))((((.(((.....)----)).)))).. ( -22.40, z-score = 1.33, R) >droVir3.scaffold_13042 3531430 92 + 5191987 CGCUGCCUCGACAACGGCAGCGGAAACUCAUAGGCAGCCGGUUGGCGUGCCGUUUGUGCAGCAUGCUCGACGGCACGACCGCC----GUUCCACAG (((((((........)))))))..........((.(((.(((.(((((((((((.(.((.....)).)))))))))).)))))----))))).... ( -42.40, z-score = -2.36, R) >droGri2.scaffold_15203 2710293 92 + 11997470 CGCUGCUGGGCAAGGGGAAGUGGGAACUCAAAGGCAUCGCGAUUCUGUGCCGCCUGAGCAGCAUGUUCGACGGCACGUCCUCC----GUUCCACAG .((((((((((..(((..........)))...(((((.(.....).))))))))).)))))).(((..(((((........))----)))..))). ( -32.00, z-score = 0.27, R) >droMoj3.scaffold_6308 755291 92 + 3356042 CGCUGUUGCGCCAAAGGCAAUGGAAACUCGAAUGCAGUCGGGUUCUUGGCAUCCCGAGCAGCAUGUUCGAUGGCCCGCCCGCC----AUUCCACAG .(((((((((((...)))...(....)......))).(((((..........)))))))))).(((..((((((......)))----)))..))). ( -30.20, z-score = -0.17, R) >triCas2.ChLG5 9887786 96 + 18847211 CUGUAUUGAGUAAGUGGGAUUGUGAAUUUAAACUGUAGCGGGUUUUUUGCACGCGAAGCGGCCAAUUCAAUUGCUUGCCCCCCUGAGAGUUUAGAG .........(((((..(.....((((((....((((.(((.((.....)).)))...))))..)))))).)..))))).................. ( -18.70, z-score = 1.35, R) >consensus CGCUGCUGGGCAAGUGGCAGCGGAAACUCGUAGGCCAGCGGAUCGCUGGCCAGCUGAGCGGCAUGCUCAAUGGCCCGUCCUCC____GUUCCACAG .((((((.....)))))).....................((((....(((((..(((((.....))))).))))).))))................ (-16.83 = -16.68 + -0.15)

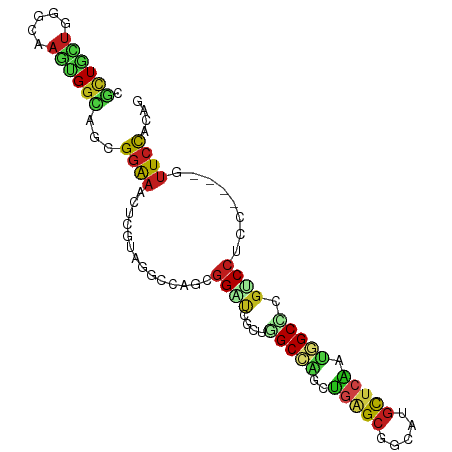
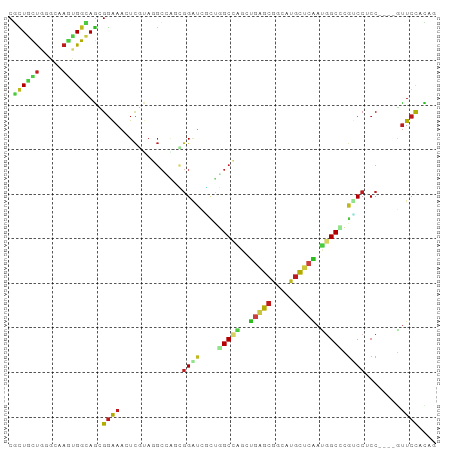
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:49 2011