| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,583,944 – 19,584,100 |
| Length | 156 |
| Max. P | 0.589910 |

| Location | 19,583,944 – 19,584,100 |
|---|---|
| Length | 156 |
| Sequences | 5 |
| Columns | 156 |
| Reading direction | reverse |
| Mean pairwise identity | 62.74 |
| Shannon entropy | 0.66965 |
| G+C content | 0.37750 |
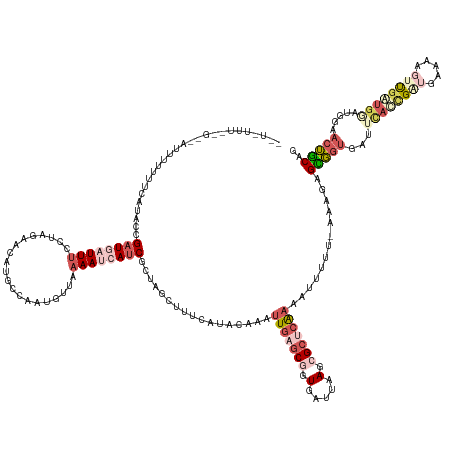
| Mean single sequence MFE | -39.24 |
| Consensus MFE | -10.99 |
| Energy contribution | -14.43 |
| Covariance contribution | 3.44 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
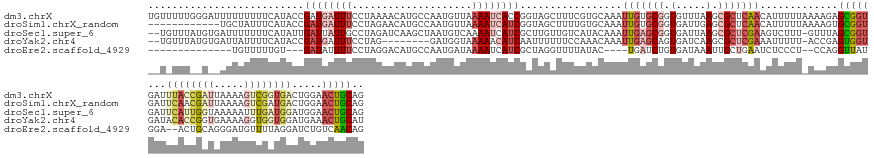
>dm3.chrX 19583944 156 - 22422827 UGUUUUUGGGAUUUUUUUUUCAUACCGAUGAUUUCCUAAAACAUGCCAAUGUUAAAAUCACCGGUAGCUUUCGUGCAAAUUGUGCGGUGUUUAAGCGCUCAACAUUUUUAAAAGAGCGGUGAUUUACCGAUUAAAAGUCGGUGACUGGAACUGCAG ((((((.((((..((....((.....)).))..))))))))))..(((..(((.((((((((....(((((((..(.....)..))).....))))((((.............))))))))))))((((((.....))))))))))))........ ( -41.32, z-score = -1.57, R) >droSim1.chrX_random 5135527 144 - 5698898 ------------UGCUAUUUCAUACCGAUGAUUUCCUAGAACAUGCCAAUGUUAAAAUCAUCGGUAGCUUUUGUGCAAAUUGUGCGGUGAUUGAGCGCUCAACAUUUUUAAAAGUGCGGUGAUUCAACGAUUAAAAGUCGAUGACUGGAACUGCAG ------------..........((((((((((((.....(((((....))))).))))))))))))(((((((......(((.(((.(.....).))).)))......)))))))(((((...(((.((((.....)))).))).....))))).. ( -40.90, z-score = -2.19, R) >droSec1.super_6 1984821 153 + 4358794 --UGUUUAUGUGAUUUUUUUCAUAUUGAUUAUUGCCUAGAUCAAGCUAAUGUCAAAAUCAUCGCUUGUUGUCAUACAAAUUGAGCGGUGAUUAAGCGCUCGAAGUCUUU-GUUUAGCGGUGAUUCAUUGGUAAAAAUUUGAUGGAUGGAACUGCAG --......(((..(((..(((((...((((.(((((..(((((.(((((...((((((((((((((.(((.....)))...)))))))))))..((.......))..))-).)))))..)))))....))))).))))..)))))..)))..))). ( -40.50, z-score = -1.73, R) >droYak2.chr4 1345287 145 + 1374474 --UGUUUAUGUGAUUAUUUUCAUACCGAUGAUUUCCUAG--------GAUGGUAAAAACAUCAAUUUUUUCCAAACAAAUUGAGCAGUGAUCAAGCGCUCGAAAUUUUU-ACCGAGUGGUGAUACACCGGUGAAAAGGUGGUGGAUGAAACUGCAU --(((((.(((((......)))))..((((.(((((...--------...))..))).))))..........)))))......(((((..(((..(((.((...(((((-((((.(((......))))))))))))..)))))..))).))))).. ( -39.00, z-score = -2.05, R) >droEre2.scaffold_4929 3669977 131 + 26641161 --------------UGUUUUUGU---GAUAUUUUCCUAGGACAUGCCAAUGAUAAAAUCAUCGCUAGGUUUUAUAC----UGAUCUGUGAUAAAUUGCUGAAUCUCCCU--CCAGGUUAUGGA--ACUGCAGGGAUGUUUUAGGAUCUGUCAACAG --------------......(((---((((...(((((((((((.((............(((((.(((((......----.)))))))))).................(--(((.....))))--......)).)))))))))))..)))).))). ( -34.50, z-score = -1.46, R) >consensus __U_UUU__G__AUUUUUUUCAUACCGAUGAUUUCCUAGAACAUGCCAAUGUUAAAAUCAUCGCUAGCUUUCAUACAAAUUGAGCGGUGAUUAAGCGCUCAAAAUUUUU_AAAGAGCGGUGAUUCACCGAUGAAAAGUUGAUGGAUGGAACUGCAG ..........................((((((((....................)))))))).................(((((((.(.....).))))))).............(((((...((((((((.....)))))))).....))))).. (-10.99 = -14.43 + 3.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:48 2011