
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,575,355 – 19,575,501 |
| Length | 146 |
| Max. P | 0.888653 |

| Location | 19,575,355 – 19,575,501 |
|---|---|
| Length | 146 |
| Sequences | 4 |
| Columns | 148 |
| Reading direction | forward |
| Mean pairwise identity | 68.95 |
| Shannon entropy | 0.50578 |
| G+C content | 0.56634 |
| Mean single sequence MFE | -48.22 |
| Consensus MFE | -25.95 |
| Energy contribution | -27.70 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19575355 146 + 22422827 GGCAACGAUUACAGCCACGCCUUGCAAUCCGAAGACGCCUGUUGUCUGCUGUAUACCUUAUGUAUACAGACUCCUGGCGAUUCUGG--GGGAAAUGGUGGGUCUGCCUCCUCCGAUUUCCGUGGAAGUGCAUUCGCCCAGAAUGCCGC (((..............((((...........(((((.....))))).((((((((.....))))))))......))))(((((((--(.(((.(((.(((......))).)))(((((....)))))...))).))))))))))).. ( -50.10, z-score = -1.33, R) >droEre2.scaffold_4690 9802310 111 + 18748788 ---------------------------------GGCUACCCACAUCUGCUUGGCACUUGAUGUGUGCC-ACGCCAUGCGCGUUGAACUAGGAAACUAGAGG---GCCUCCUCCGAUUUCCGUGGAAGUGGACUCGCCCAGAACGCCGU ---------------------------------((....))......((.((((((.......)))))-).)).(((.(((((...((((....)))).((---((....(((.(((((....))))))))...))))..)))))))) ( -41.40, z-score = -2.00, R) >droSec1.super_8 1854910 144 + 3762037 GGC--UGAUUACAGUCACGCCUUGCAAUCCGAAGACGCCUGUCGUCCAUUGUAUACCCUAUGUAUACAGACUCCUAGCGAGUCUGG--GGGCAAUGGAGGGCCUGCCUCCUCCGAUUUCCGUGGAAGUGCAUUCGCCCAGAAUGCCGC (((--((((....)))).)))..((((((.((.((.((..(((.((((((((...(((...(....)((((((.....))))))))--).)))))))).)))..)).)).)).)))).........(.((((((.....))))))))) ( -50.70, z-score = -1.50, R) >droSim1.chrX_random 5128952 137 + 5698898 GGC--UGAUUACAGCCACGCCUUGCAAUGCGAAGACGCCU-------AUUGUAUACCCUAUGUAUACAGACUCCUGGCGAGUCUGG--GGGCAAUGGAGGGCCUGCCUCCUCCGAUUUCCGUGGAAGUGCAUUCGCCCAGAAUGCCGC (((--((....)))))..(((((((((((((....)))..-------)))))..............(((((((.....))))))))--))))..(((((((......)))))))(((((....)))))((((((.....))))))... ( -50.70, z-score = -1.57, R) >consensus GGC__UGAUUACAGCCACGCCUUGCAAUCCGAAGACGCCUGUCGUCUACUGUAUACCCUAUGUAUACAGACUCCUGGCGAGUCUGG__GGGAAAUGGAGGGCCUGCCUCCUCCGAUUUCCGUGGAAGUGCAUUCGCCCAGAAUGCCGC ............................................(((((.((((((.....))))))((((((.....)))))).....((((((((((((......))))))).)))))))))).(.((((((.....))))))).. (-25.95 = -27.70 + 1.75)

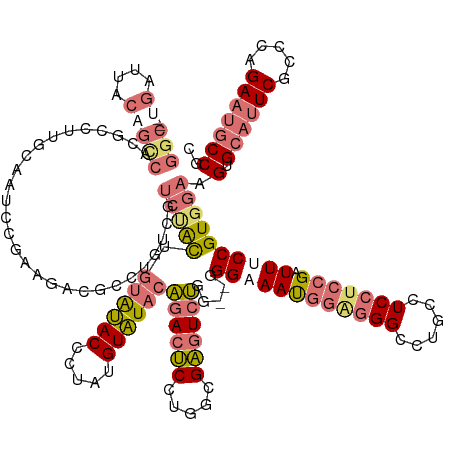

| Location | 19,575,355 – 19,575,501 |
|---|---|
| Length | 146 |
| Sequences | 4 |
| Columns | 148 |
| Reading direction | reverse |
| Mean pairwise identity | 68.95 |
| Shannon entropy | 0.50578 |
| G+C content | 0.56634 |
| Mean single sequence MFE | -49.73 |
| Consensus MFE | -27.95 |
| Energy contribution | -30.57 |
| Covariance contribution | 2.62 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.872003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19575355 146 - 22422827 GCGGCAUUCUGGGCGAAUGCACUUCCACGGAAAUCGGAGGAGGCAGACCCACCAUUUCCC--CCAGAAUCGCCAGGAGUCUGUAUACAUAAGGUAUACAGCAGACAACAGGCGUCUUCGGAUUGCAAGGCGUGGCUGUAAUCGUUGCC (((((((((((((.((((((.((((..((.....))..)))))))(......)..))).)--))))))).((((.(.(.((((((((.....))))))))).........(((((....)).)))....).)))).......))))). ( -50.10, z-score = -1.28, R) >droEre2.scaffold_4690 9802310 111 - 18748788 ACGGCGUUCUGGGCGAGUCCACUUCCACGGAAAUCGGAGGAGGC---CCUCUAGUUUCCUAGUUCAACGCGCAUGGCGU-GGCACACAUCAAGUGCCAAGCAGAUGUGGGUAGCC--------------------------------- ..(((....((((((((....)))....((((((..((((....---))))..))))))..)))))((.(((((.((.(-(((((.......)))))).))..))))).)).)))--------------------------------- ( -44.30, z-score = -2.04, R) >droSec1.super_8 1854910 144 - 3762037 GCGGCAUUCUGGGCGAAUGCACUUCCACGGAAAUCGGAGGAGGCAGGCCCUCCAUUGCCC--CCAGACUCGCUAGGAGUCUGUAUACAUAGGGUAUACAAUGGACGACAGGCGUCUUCGGAUUGCAAGGCGUGACUGUAAUCA--GCC ..(((.....((((...(((.((((..((.....))..))))))).))))(((((((...--.(((((((.....)))))))(((((.....))))))))))))..(((((((((((........)))))))..)))).....--))) ( -51.90, z-score = -1.04, R) >droSim1.chrX_random 5128952 137 - 5698898 GCGGCAUUCUGGGCGAAUGCACUUCCACGGAAAUCGGAGGAGGCAGGCCCUCCAUUGCCC--CCAGACUCGCCAGGAGUCUGUAUACAUAGGGUAUACAAU-------AGGCGUCUUCGCAUUGCAAGGCGUGGCUGUAAUCA--GCC ..((((....((((...(((.((((..((.....))..))))))).)))).....)))).--..((((((.....))))))((((((.....))))))...-------..(((((((.((...)))))))))(((((....))--))) ( -52.60, z-score = -1.57, R) >consensus GCGGCAUUCUGGGCGAAUGCACUUCCACGGAAAUCGGAGGAGGCAGGCCCUCCAUUGCCC__CCAGACUCGCCAGGAGUCUGUAUACAUAAGGUAUACAACAGACGACAGGCGUCUUCGGAUUGCAAGGCGUGGCUGUAAUCA__GCC ..((((....((((...(((.((((..((.....))..))))))).)))).....)))).....((((((.....))))))((((((.....))))))............(((((((........)))))))................ (-27.95 = -30.57 + 2.62)

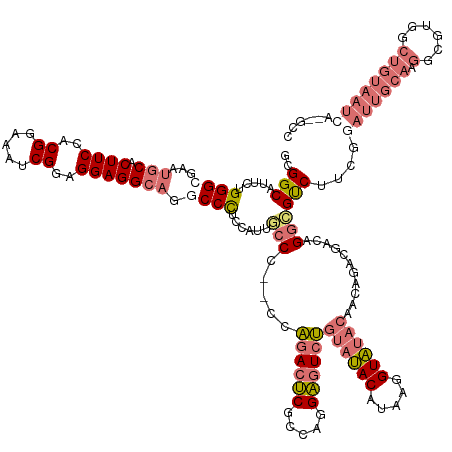
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:46 2011