
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,559,894 – 19,560,041 |
| Length | 147 |
| Max. P | 0.995763 |

| Location | 19,559,894 – 19,560,041 |
|---|---|
| Length | 147 |
| Sequences | 5 |
| Columns | 153 |
| Reading direction | forward |
| Mean pairwise identity | 71.68 |
| Shannon entropy | 0.50266 |
| G+C content | 0.54262 |
| Mean single sequence MFE | -57.98 |
| Consensus MFE | -24.42 |
| Energy contribution | -28.94 |
| Covariance contribution | 4.52 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
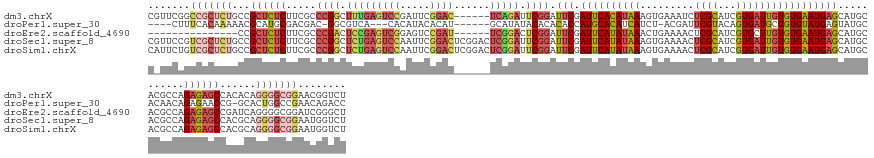
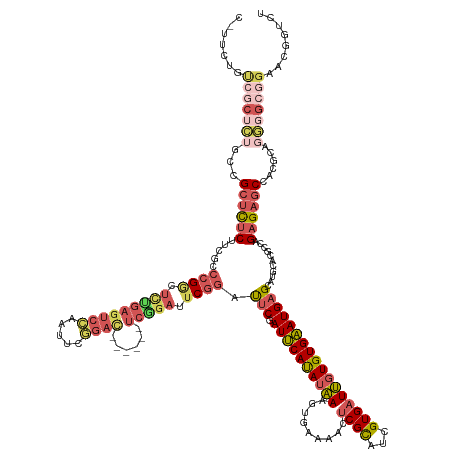
>dm3.chrX 19559894 147 + 22422827 CGUUCGGCCGCUCUGCCGCUCUCUUCGCCCGGCUUUGAGUCCGAUUCGGAC------UCAGAUUCGGAUUCGAUUCACAUAAAGUGAAAUCUCGCAUCGUGAUUGUGUGAAUGAGCAUGCACGCCAGAGAGCCACACAGGGGCGGAACGGUCU (((((.(((.((.((..(((((((..(((((..((((((((((...)))))------)))))..)))....(((((((.....))).))))..))..((((...((((......)))).))))..)))))))..)).)).))).))))).... ( -63.00, z-score = -3.33, R) >droPer1.super_30 682195 137 - 958394 ----CUUUCACAAAAACGCAUGCGACGAC-GGCGUCA---CACAUACACAU------GCAUAUACACACACCAUGCACAUCAUCU-ACGAUUCGUACAGUGAUGCUGUGUAUGAGUAUGCACAACAGAGAACCG-GCACUGGCCGAACAGACC ----.............((((((((((..-..)))).---..(((((((((------((((...........)))))((((((.(-(((...))))..)))))).)))))))).))))))............((-((....))))........ ( -39.90, z-score = -2.65, R) >droEre2.scaffold_4690 9788037 132 + 18748788 ---------------CCGCUCUCUUCGCCCGACUCCGAGUCGGAGUCCGAU------UCGGACUCGGAUUCGAUUCAUAUAAACUGAAAACUCGCAUCGUGCUUGUGUGAAUGAGCAUGCACGCCAGAGAGCCGAUCAGGGGCGGAUCGGGCU ---------------..((((..(((((((.....((((((.(((((((..------.))))))).))))))...........((((...(((....(((((.(((........))).))))).....)))....)))))))))))..)))). ( -56.60, z-score = -2.89, R) >droSec1.super_8 1840544 153 + 3762037 CGUUCCGUCGCUCUGCCGCUCUCUUCGCCCGGCUCUGAGUCCAAUUCGGACUCGGACUCGGAUUCGGAUUCGAUUCAUAUAAAGUGAAAACUCGCAUCGUGAUUGUGUGAAUGAGCAUGCACGCCAGAGAGCCACGCAGGGGCGGAAUGGUCU (((((((((.(.((((.((((((((((((((..(((((((((.....)))))))))..))).....(((.((((((((.....)))))...))).)))))))..(((((.((....)).))))).)))))))...)))))))))))))).... ( -66.10, z-score = -3.48, R) >droSim1.chrX 15082212 153 + 17042790 CAUUCUGUCGCUCUGCCGCUCUCUUCGCCCGGCUCUGAGUCCAAUUCGGACUCGGACUCGGAUUCGGAUUCGAUUCAUAUAAAGUGAAAACUCGCAUCGUGAUUGUGUGAAUGAGCAUGCACGCCAGAGAGCCACGCAGGGGCGGAAUGGUCU (((((((((.(.((((.((((((((((((((..(((((((((.....)))))))))..))).....(((.((((((((.....)))))...))).)))))))..(((((.((....)).))))).)))))))...)))))))))))))).... ( -64.30, z-score = -3.42, R) >consensus C_UUCUGUCGCUCUGCCGCUCUCUUCGCCCGGCUCUGAGUCCAAUUCGGAC______UCGGAUUCGGAUUCGAUUCAUAUAAAGUGAAAACUCGCAUCGUGAUUGUGUGAAUGAGCAUGCACGCCAGAGAGCCACGCAGGGGCGGAACGGUCU (((((.(((.((.....((((((.....((((.(((((((((.....))))......))))).)))).(((.((((((((((.........((((...)))))))))))))))))...........)))))).....)).))).))))).... (-24.42 = -28.94 + 4.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:41 2011