| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,011,495 – 12,011,569 |
| Length | 74 |
| Max. P | 0.840098 |

| Location | 12,011,495 – 12,011,569 |
|---|---|
| Length | 74 |
| Sequences | 12 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 93.55 |
| Shannon entropy | 0.13749 |
| G+C content | 0.42446 |
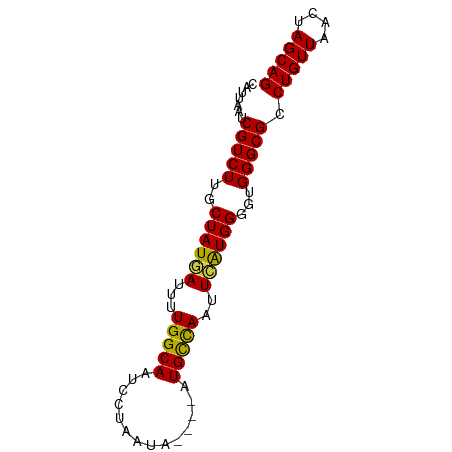
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -18.09 |
| Energy contribution | -17.91 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.19 |
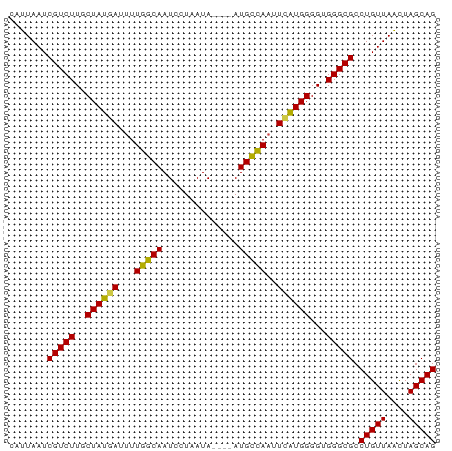
| Mean z-score | -1.74 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
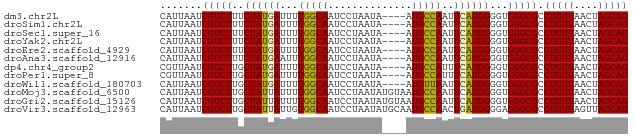
>dm3.chr2L 12011495 74 + 23011544 CAUUAAUCGUCUUUCUAUGAUUUUGGCAAUCCUAAUA----AUGCCAAUUCAUGGGGUGGGCGCCUGUUAACUAGCAG .......(((((((((((((..((((((.........----.)))))).)))))))).))))).(((((....))))) ( -21.80, z-score = -2.68, R) >droSim1.chr2L 11815588 74 + 22036055 CAUUAAUCGUCUUUCUAUGAUUUUGGCAAUCCUAAUA----AUGCCAAUUCAUGGGGUGGGCGCCUGUUAACUAGCAG .......(((((((((((((..((((((.........----.)))))).)))))))).))))).(((((....))))) ( -21.80, z-score = -2.68, R) >droSec1.super_16 207064 74 + 1878335 CAUUAAUCGUCUUUCUAUGAUUUUGGCAAUCCUAAUA----AUGCCAAUUCAUGGGGUGGGCGCCUGUUAACUAGCAG .......(((((((((((((..((((((.........----.)))))).)))))))).))))).(((((....))))) ( -21.80, z-score = -2.68, R) >droYak2.chr2L 8430740 74 + 22324452 CAUUAAUCGUCUUUCUAUGAUUUUGGCAAUCCUAAUA----AUGCCAAUUCAUGGGGUGGGCGCCUGUUAACUAGCAG .......(((((((((((((..((((((.........----.)))))).)))))))).))))).(((((....))))) ( -21.80, z-score = -2.68, R) >droEre2.scaffold_4929 13238990 74 - 26641161 CAUUAAUCGUCUUUCUAUGAUUUUGGCAAUCCUAAUA----AUGCCAAUUCAUGGGGUGGGCGCCUGUUAACUAGCAG .......(((((((((((((..((((((.........----.)))))).)))))))).))))).(((((....))))) ( -21.80, z-score = -2.68, R) >droAna3.scaffold_12916 6724616 74 - 16180835 CAUUAAUCGUCUUUCUAUGAAUUUGGCAAUCCUAAUA----AUGCCAAUUCGUGGGGUGGGCGCCUGUUAACUAGCAG .......((((((((((((((((.((((.........----.))))))))))))))).))))).(((((....))))) ( -22.30, z-score = -2.79, R) >dp4.chr4_group2 289159 74 - 1235136 CGUUAAUCGUCUUGCUAUGAUUUUGGCAAUCCUAAUA----AUGCCAUUUCAUGGGGUGGGCGCCUGUUAACUAGCAG .((((((((((((.((((((...(((((.........----.)))))..)))))).).)))))...))))))...... ( -21.80, z-score = -1.94, R) >droPer1.super_8 2950120 74 + 3966273 CGUUAAUCGUCUUGCUAUGAUUUUGGCAAUCCUAAUA----AUGCCAUUUCAUGGGGUGGGCGCCUGUUAACUAGCAG .((((((((((((.((((((...(((((.........----.)))))..)))))).).)))))...))))))...... ( -21.80, z-score = -1.94, R) >droWil1.scaffold_180703 1188663 74 - 3946847 CAUUAAUCGUCUUGCUAUGAUUUUGGCAAUCCUAAUA----AUGUUAAUUCAUGGGGUGGGCGCCUGUUAACUAGCAG .......((((((.((((((..((((((.........----.)))))).)))))).).))))).(((((....))))) ( -16.00, z-score = -0.54, R) >droMoj3.scaffold_6500 26590669 78 - 32352404 CAUUAAUCGUCUUGCUAUUAUUUUGGCAAUCCUAAUAUGUAAUGCCAAUUCAUGGGGUGGGCGCCUGUUAACUAGCAG ............((((((((....(((...((((.((((.(((....)))))))...)))).)))...))).))))). ( -16.00, z-score = 0.18, R) >droGri2.scaffold_15126 2396646 78 - 8399593 CAUUAAUCGUCUUGCUAUUAUUUUGGCAAUCCUAAUAUGUAAUGCCAAUUCAUGGGGUGGGCGCCUGUUAACUAGCAG ............((((((((....(((...((((.((((.(((....)))))))...)))).)))...))).))))). ( -16.00, z-score = 0.18, R) >droVir3.scaffold_12963 13888455 78 + 20206255 CAUUAAUCGUCUUGCUAUUAUUGUGGCAAUCCUAAUAUGCAAUGCCAACUGAUGGGGAGGGCGCCUGUUAGUUAGCAG ........(..(((((((....)))))))..)..........(((.((((((((((.......)))))))))).))). ( -21.50, z-score = -0.69, R) >consensus CAUUAAUCGUCUUGCUAUGAUUUUGGCAAUCCUAAUA____AUGCCAAUUCAUGGGGUGGGCGCCUGUUAACUAGCAG .......(((((..((((((...(((((..............)))))..))))))...))))).(((((....))))) (-18.09 = -17.91 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:15 2011