
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,469,618 – 19,469,723 |
| Length | 105 |
| Max. P | 0.939031 |

| Location | 19,469,618 – 19,469,723 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 61.68 |
| Shannon entropy | 0.75436 |
| G+C content | 0.69485 |
| Mean single sequence MFE | -49.52 |
| Consensus MFE | -12.28 |
| Energy contribution | -11.99 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.76 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
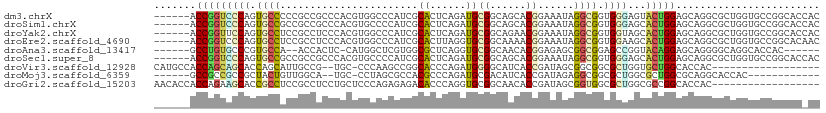
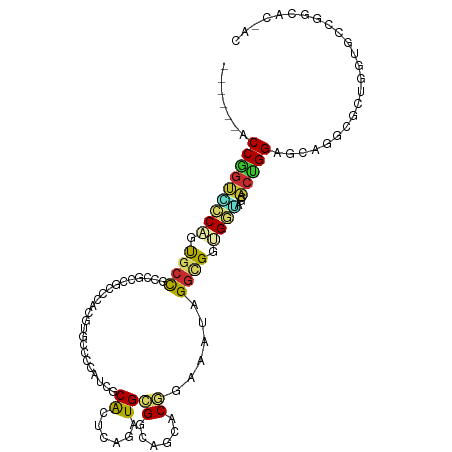
>dm3.chrX 19469618 105 + 22422827 ------ACCGGUCCCAGUGCCCCCGCCGCCCACGUGGCCCAUCGCACUCAGAUGCGGCAGCACGGAAAUAGGCGGUGGGAGUACUGGAGCAGGCGCUGGUGCCGGCACCAC ------.((((..(((((((((((((((((..(((((((((((.......)))).)))..))))......))))))))).((......)).))))))))..))))...... ( -58.90, z-score = -3.41, R) >droSim1.chrX 15034321 105 + 17042790 ------ACCGGUCCCAGUGCCGCCGCCGCCCACGUGCCCCAUCGCACUCAGAUGCGGCAGCACGGAAAUAGGCGGUGGGAGCACUGGAGCAGGCGCUGGUGCCGGCACCAC ------.((((..((((((((.((((((((..(((((((((((.......)))).))..)))))......))))))))..((......)).))))))))..))))...... ( -58.50, z-score = -3.17, R) >droYak2.chrX 9901484 105 - 21770863 ------ACCGGUUCCAGUGCCUCCGCCUCCCACGUGGCCCAUCGCACUCAGAUGCGGCAGAACGGAAAUAGGCGGUGGUAGCACUGGAGCAGGCGCUGGUGCCGGCACCAC ------....((((((((((..(((((.((..(((.(((((((.......)))).)))...)))......)).)))))..)))))))))).((.((((....)))).)).. ( -54.50, z-score = -3.25, R) >droEre2.scaffold_4690 9705432 105 + 18748788 ------ACCGGUCCCAGUGCCUCCGCCUCCCACGUGGCCCAUCGCACUUAGGUGCGGCAAAACGGAAAUAGGCGGUGGAAGCACUGGAGCAGGCGCUGGUGCCGGCACAAC ------.((((..(((((((((((((((....(((.(((((((.......)))).)))...))).....))))((((....)))))))...))))))))..))))...... ( -52.40, z-score = -2.65, R) >droAna3.scaffold_13417 4514961 96 - 6960332 ------GCCUGUGCCCGUGCCA--ACCACUC-CAUGGCUCGUGGCGCUCAGGUGCGGCAACACGGAGAGCGGCGGAGCCGGUACAGGAGCAGGGGCAGGCACCAC------ ------((((..((((.(((..--.((....-..((((((.((.(((((...((.(....).))..))))).)))))))).....)).))).)))))))).....------ ( -44.40, z-score = -0.57, R) >droSec1.super_8 1752240 105 + 3762037 ------ACCGGUCCCAGUGCCGCCGCCGCCCACGUGCCCCAUCGCACUCAGAUGCGGCAGCACGGAAAUAGGCGGUGGGAGCACUGGAGCAGGCGCUGGUGCCGGCACCAC ------.((((..((((((((.((((((((..(((((((((((.......)))).))..)))))......))))))))..((......)).))))))))..))))...... ( -58.50, z-score = -3.17, R) >droVir3.scaffold_12928 2093243 90 - 7717345 CAUGCCACCAGCAGCACCAGCAUUGCCG--UGC-CCCAAGCCGGCACCCAGAUGGGGCAUCACCGAUAGCGGCGGCGCUGGUGCUGGCACCAC------------------ ..(((((......((((((((.((((((--(((-((((..(.((...)).).))))))(((...))).))))))).)))))))))))))....------------------ ( -43.40, z-score = -2.08, R) >droMoj3.scaffold_6359 377353 90 - 4525533 ------GCCGCCGCCGCUACUGUUGGCA--UGC-CCUAGCGCCACGCCCAGAUGCGACAUCACCGAUAGAGGCGGCGCUGGCGCUGGCGCAGGCACCAC------------ ------...((((((((....).)))).--(((-.(((((((((((((..((((...)))).((......)).)))).))))))))).)))))).....------------ ( -43.40, z-score = -1.88, R) >droGri2.scaffold_15203 10489650 93 - 11997470 AACACCACCAGAAGCACCGCCUCCGCCUCCUGCUCCCAGAGAGACACCCAGGUGCGGCAACACCGAUAGCGGUGGCGCUGGCGCCGGCACCAC------------------ ..................(((...(.((((((....))).))).).....((((((((..(((((....)))))..))).)))))))).....------------------ ( -31.70, z-score = -0.50, R) >consensus ______ACCGGUCCCAGUGCCGCCGCCGCCCACGUGCCCCAUCGCACUCAGAUGCGGCAGCACGGAAAUAGGCGGUGGUAGCACUGGAGCAGGCGCUGGUGCCGGCAC_AC .......(((((((((.((((.......................((......))((......))......)))).))))...)))))........................ (-12.28 = -11.99 + -0.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:29 2011