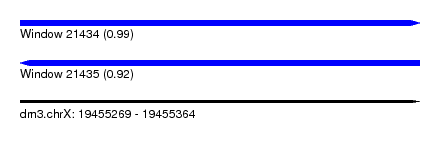
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,455,269 – 19,455,364 |
| Length | 95 |
| Max. P | 0.991674 |

| Location | 19,455,269 – 19,455,364 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 71.84 |
| Shannon entropy | 0.55896 |
| G+C content | 0.51769 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -18.73 |
| Energy contribution | -19.86 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
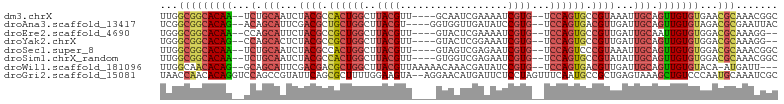
>dm3.chrX 19455269 95 + 22422827 UUGGCGGCACAA--UCUGCAAUCUACGCCACUGGCUUACGUU----GCAAUCGAAAAUCGUG--UCCAGUGCCGUAAAUUGCAGUUGUGUGAACGCAAACGGC ...(((((((((--.(((((((.((((.((((((..((((..----............))))--.)))))).)))).)))))))))))))...)))....... ( -37.44, z-score = -3.39, R) >droAna3.scaffold_13417 4499716 95 - 6960332 UCGGCGGCACAG--ACAGCAUUCGACGCUGCUGGCUUACGU----GGUGGUUGAUAUCCGUG--UCCAGUGACGUUGAUUGCAGUUGUGUAGACGCGAAUUAC ...(((((((((--...(((.((((((..(((((..((((.----((((.....))))))))--.)))))..)))))).)))..))))))...)))....... ( -37.30, z-score = -2.07, R) >droEre2.scaffold_4690 9691198 93 + 18748788 UGGGCGGCACAA--CCAGCAUUCUACGCCGCUGGCUUACGUU----GUACUCGAAAAUCGUG--UCCAGUGCCGUUGAUUGCAAUUGUGUGGACGCAAAGG-- ((.((.((((((--...(((.((.(((.((((((..((((..----............))))--.)))))).))).)).)))..)))))).).).))....-- ( -30.84, z-score = -0.62, R) >droYak2.chrX 9887014 93 - 21770863 UGGGCGGCACAG--CCAGCACUCUACGCCGCUGGCUUACGUU----GUACUCGGAAAUCGUG--UCCAGUGCCGUUGAUUGCAGUUGUGUGGACGCAAAGG-- ((.((.((((((--(..(((.((.(((.((((((..((((..----............))))--.)))))).))).)).))).))))))).).).))....-- ( -33.44, z-score = -0.24, R) >droSec1.super_8 1737865 95 + 3762037 UUGGCGGCACAA--UCUGCAAUCUACGCCACUGGCUUACGUU----GUAGUCGAGAAUCGUG--UCCAGUCCCGUAAAUUGCAGUUGUGUGGACGCAAACGGC ...(((((((((--.(((((((.((((..(((((..((((((----.(.....).)).))))--.)))))..)))).)))))))))))))...)))....... ( -33.90, z-score = -1.97, R) >droSim1.chrX_random 5079007 95 + 5698898 UUGGCGGCACAA--UCUGCAAUCUACGCCACUGGCUUACGUU----GUGGUCGAGAAUCGUG--UCCAGUGCCGUAUAUUGCAGUUGUGUGGACGCAAACGGC ...(((((((((--.(((((((.((((.((((((..((((((----.(.....).)).))))--.)))))).)))).)))))))))))))...)))....... ( -37.60, z-score = -2.22, R) >droWil1.scaffold_181096 7961114 95 + 12416693 UUGGCAACACAG--GCAGCAUUCGACGACGCUGGCUUACGUUAAAAACAAACGAUAUCCGUG--UCCAGUGACGUUGAUUGCAGUUGUGUACA-AUGAUU--- (((...((((((--.(.(((.((((((.((((((..((((..................))))--.)))))).)))))).))).))))))).))-).....--- ( -29.57, z-score = -1.87, R) >droGri2.scaffold_15081 2233665 101 - 4274704 UAACCAACACAGGUCCAGCCGUAUUCAGCGCUUUUGGAAGUA--AGGAACAUGAUUCUCCUAGUUUCAAUGCCGCUGAGUAAAGCUGUCCCAAUGCAAAUCGC ...........((..((((..(((((((((...((((((.((--.(((.........))))).))))))...)))))))))..))))..))............ ( -33.30, z-score = -4.13, R) >consensus UUGGCGGCACAA__CCAGCAAUCUACGCCGCUGGCUUACGUU____GUACUCGAAAAUCGUG__UCCAGUGCCGUUGAUUGCAGUUGUGUGGACGCAAACGGC ...(((((((((.....(((...((((.((((((..((((..................))))...)))))).))))...)))..))))))...)))....... (-18.73 = -19.86 + 1.13)

| Location | 19,455,269 – 19,455,364 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 71.84 |
| Shannon entropy | 0.55896 |
| G+C content | 0.51769 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -14.46 |
| Energy contribution | -15.37 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
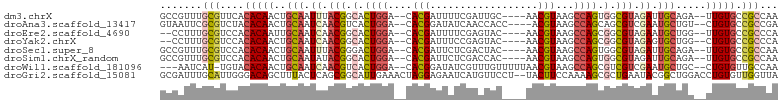
>dm3.chrX 19455269 95 - 22422827 GCCGUUUGCGUUCACACAACUGCAAUUUACGGCACUGGA--CACGAUUUUCGAUUGC----AACGUAAGCCAGUGGCGUAGAUUGCAGA--UUGUGCCGCCAA .......(((....(((((((((((((((((.((((((.--.(((............----..)))...)))))).)))))))))))).--))))).)))... ( -39.34, z-score = -4.24, R) >droAna3.scaffold_13417 4499716 95 + 6960332 GUAAUUCGCGUCUACACAACUGCAAUCAACGUCACUGGA--CACGGAUAUCAACCACC----ACGUAAGCCAGCAGCGUCGAAUGCUGU--CUGUGCCGCCGA .......(((....((((((.(((.((.((((..((((.--.(((.............----.)))...))))..)))).)).))).))--.)))).)))... ( -25.44, z-score = -0.58, R) >droEre2.scaffold_4690 9691198 93 - 18748788 --CCUUUGCGUCCACACAAUUGCAAUCAACGGCACUGGA--CACGAUUUUCGAGUAC----AACGUAAGCCAGCGGCGUAGAAUGCUGG--UUGUGCCGCCCA --.....(((....((((((((((.((.(((.(.(((((--(.((.....)).))..----..(....))))).).))).)).))).))--))))).)))... ( -27.30, z-score = -0.37, R) >droYak2.chrX 9887014 93 + 21770863 --CCUUUGCGUCCACACAACUGCAAUCAACGGCACUGGA--CACGAUUUCCGAGUAC----AACGUAAGCCAGCGGCGUAGAGUGCUGG--CUGUGCCGCCCA --...(((((..........)))))....((((((((((--.......)))).....----......((((((((.(.....)))))))--)))))))).... ( -28.20, z-score = 0.04, R) >droSec1.super_8 1737865 95 - 3762037 GCCGUUUGCGUCCACACAACUGCAAUUUACGGGACUGGA--CACGAUUCUCGACUAC----AACGUAAGCCAGUGGCGUAGAUUGCAGA--UUGUGCCGCCAA .......(((....(((((((((((((((((..(((((.--.(((............----..)))...)))))..)))))))))))).--))))).)))... ( -34.04, z-score = -2.74, R) >droSim1.chrX_random 5079007 95 - 5698898 GCCGUUUGCGUCCACACAACUGCAAUAUACGGCACUGGA--CACGAUUCUCGACCAC----AACGUAAGCCAGUGGCGUAGAUUGCAGA--UUGUGCCGCCAA .......(((....((((((((((((.((((.((((((.--.(((............----..)))...)))))).)))).))))))).--))))).)))... ( -35.74, z-score = -3.31, R) >droWil1.scaffold_181096 7961114 95 - 12416693 ---AAUCAU-UGUACACAACUGCAAUCAACGUCACUGGA--CACGGAUAUCGUUUGUUUUUAACGUAAGCCAGCGUCGUCGAAUGCUGC--CUGUGUUGCCAA ---......-.(((((((...(((.((.(((.(.((((.--.(((..................)))...)))).).))).)).)))...--.)))).)))... ( -22.67, z-score = -0.02, R) >droGri2.scaffold_15081 2233665 101 + 4274704 GCGAUUUGCAUUGGGACAGCUUUACUCAGCGGCAUUGAAACUAGGAGAAUCAUGUUCCU--UACUUCCAAAAGCGCUGAAUACGGCUGGACCUGUGUUGGUUA ..(((..((((.((..(((((.((.((((((...(((.((.((((.((((...))))))--)).)).)))...)))))).)).)))))..)).))))..))). ( -33.50, z-score = -2.61, R) >consensus GCCAUUUGCGUCCACACAACUGCAAUCAACGGCACUGGA__CACGAUUAUCGACUAC____AACGUAAGCCAGCGGCGUAGAAUGCUGA__CUGUGCCGCCAA .......(((....(((((..(((.((.(((.(.((((....(((..................)))...)))).).))).)).))).....))))).)))... (-14.46 = -15.37 + 0.91)
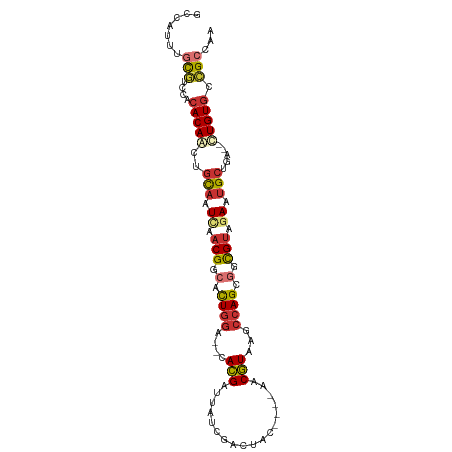
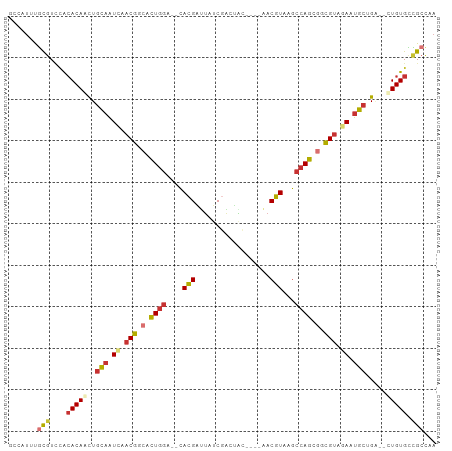
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:27 2011