| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,452,934 – 19,453,035 |
| Length | 101 |
| Max. P | 0.999940 |

| Location | 19,452,934 – 19,453,035 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.63 |
| Shannon entropy | 0.65452 |
| G+C content | 0.49387 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -21.21 |
| Energy contribution | -20.36 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.43 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.05 |
| SVM RNA-class probability | 0.999940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
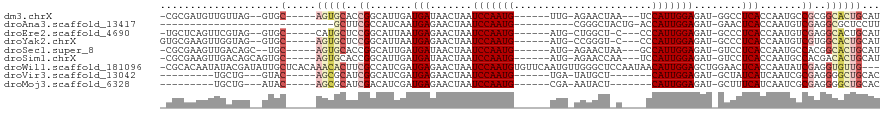
>dm3.chrX 19452934 101 + 22422827 AUGCAGUGCCGCGGCAUUGGUGAGGCC-AUCUCCAAUGGA---UUAGUUCU-CAA------CAUUGGAUUAGUUAUCAUCAAUGCCGGUGCACU-----GCAC--CUAACAACAUCGCG- .(((((((((.((((((((((((....-...(((((((..---........-...------))))))).......))))))))))))).)))))-----))).--..............- ( -43.76, z-score = -5.26, R) >droAna3.scaffold_13417 4492670 79 - 6960332 AAGGAGCGCCUCGACAUUGGUGAGUUC-AUCUCCAAUGGU-CAGUAGCCCG----------CAUUGGAUUAGUUCUCAUUGAUGGCGAAGC----------------------------- .......((.(((.(((..(((((..(-...(((((((..-..........----------)))))))...)..)))))..))).))).))----------------------------- ( -25.50, z-score = -1.53, R) >droEre2.scaffold_4690 9688913 100 + 18748788 AUGCAGUGCCUCGACAUUGGUGAGGGC-AUCUCCAAUGGG---G-AGCCAG-CAU------CAUUGGAUUAGUUCUCAUUAAUGCCGGAGCAUG-----GCAC--CUACGAACUGAGCA- .(((.((((..((.(((((((((((((-...(((((((..---(-......-)..------)))))))...))))))))))))).))..)))).-----))).--..............- ( -38.90, z-score = -2.32, R) >droYak2.chrX 9884962 101 - 21770863 AUGCAGUGCCACGACAUUGGUGAGGGC-AUCUCCAAUGGG---G-ACCCGG-CAU------CAUUGGAUUAGUUCUCAUUAAUGCCGGAGCACU-----GCAC--CUACCAACUUCGCAC .((((((((..((.(((((((((((((-...(((((((..---(-......-)..------)))))))...))))))))))))).))..)))))-----))).--............... ( -44.20, z-score = -4.73, R) >droSec1.super_8 1735244 101 + 3762037 AUGCAGUGCCGUGGCAUUGGUGAGGAC-AUCUCCAAUGGC---UUAGUUCU-CAU------CAUUGGAUUAGUUAUCAUCAAUGCCGGUGCACU-----GCA--GCUGUCAACUUCGCG- .((((((((..((((((((((((.(((-...((((((((.---........-..)------)))))))...))).))))))))))))..)))))-----)))--((..........)).- ( -43.90, z-score = -4.80, R) >droSim1.chrX 15018510 103 + 17042790 AUGCAGUGUCGUGGCAUUGGUGAGGAC-AUCUCCAAUGGA---UUGGUUCU-CAU------CAUUGGAUUAGUUAUCAUCAAUGCCGGUGCACU-----GCACUGCUGUCAACUUCGCG- .((((((((..((((((((((((.(((-...((((((((.---.((.....-)))------)))))))...))).))))))))))))..)))))-----))).................- ( -42.50, z-score = -4.13, R) >droWil1.scaffold_181096 7956783 116 + 12416693 ---CAACACCUCGAUAUUGGUGAGUUCCAGCUCCAAUGUUAUUGGAGCCCAACAUUGAACACAUUGGAUUAGUUCUCAUCGAUGGCGAAGUGUUUGUGAGCAAUAUCGUAUAUUGUGCG- ---.(((((.(((.((((((((((..(..((((((((...))))))))((((...........))))....)..)))))))))).))).)))))...(.(((((((...))))))).).- ( -35.80, z-score = -2.52, R) >droVir3.scaffold_13042 4285165 88 - 5191987 GUGCAGCCCCUCGCGAUUGAUGAUAGC-AUCUCCAAUG-------AGCAUA-UCA------CAUUGGAUUAGUUCUCAUCGAUGCCGAUGCGCU-----GUAC---CAGCA--------- (((((((.(.(((..((((((((.(((-...(((((((-------......-...------)))))))...))).))))))))..))).).)))-----))))---.....--------- ( -32.60, z-score = -3.37, R) >droMoj3.scaffold_6328 1552363 88 - 4453435 GUGCAGCCCCUCGCGAUUGAUGAAAGC-AUCUCCAAUG-------AGUAUU-UCG------CAUUGGAUUAGUUCUCAUCGAUGUCGAUGCGCU-----GUAU---CAGCA--------- (((((((.(.(((..((((((((.(((-...(((((((-------.(....-.).------)))))))...))).))))))))..))).).)))-----))))---.....--------- ( -31.80, z-score = -3.07, R) >consensus AUGCAGUGCCUCGACAUUGGUGAGGGC_AUCUCCAAUGG____G_AGCCCU_CAU______CAUUGGAUUAGUUCUCAUCAAUGCCGGUGCACU_____GCAC__CUAUCAACUUCGCG_ ....(((((..((.(((((((((........(((((((.......................))))))).......))))))))).))..))))).......................... (-21.21 = -20.36 + -0.85)



| Location | 19,452,934 – 19,453,035 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.63 |
| Shannon entropy | 0.65452 |
| G+C content | 0.49387 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -9.44 |
| Energy contribution | -9.89 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19452934 101 - 22422827 -CGCGAUGUUGUUAG--GUGC-----AGUGCACCGGCAUUGAUGAUAACUAAUCCAAUG------UUG-AGAACUAA---UCCAUUGGAGAU-GGCCUCACCAAUGCCGCGGCACUGCAU -..............--((((-----(((((..((((((((.(((...(((.(((((((------..(-(.......---)))))))))..)-))..))).))))))))..))))))))) ( -42.30, z-score = -4.43, R) >droAna3.scaffold_13417 4492670 79 + 6960332 -----------------------------GCUUCGCCAUCAAUGAGAACUAAUCCAAUG----------CGGGCUACUG-ACCAUUGGAGAU-GAACUCACCAAUGUCGAGGCGCUCCUU -----------------------------((((((.(((...((((......(((((((----------(((....)))-..)))))))...-...))))...))).))))))....... ( -25.70, z-score = -2.94, R) >droEre2.scaffold_4690 9688913 100 - 18748788 -UGCUCAGUUCGUAG--GUGC-----CAUGCUCCGGCAUUAAUGAGAACUAAUCCAAUG------AUG-CUGGCU-C---CCCAUUGGAGAU-GCCCUCACCAAUGUCGAGGCACUGCAU -..........((((--.(((-----(.....(((((((((.((.((.....)))).))------)))-)))).(-(---..((((((.((.-....)).))))))..)))))))))).. ( -34.00, z-score = -1.78, R) >droYak2.chrX 9884962 101 + 21770863 GUGCGAAGUUGGUAG--GUGC-----AGUGCUCCGGCAUUAAUGAGAACUAAUCCAAUG------AUG-CCGGGU-C---CCCAUUGGAGAU-GCCCUCACCAAUGUCGUGGCACUGCAU ((((.(.((((((((--(.((-----(...(((((((((((.((.((.....)))).))------)))-))((..-.---.))...)))).)-))))).))))))....).))))..... ( -42.70, z-score = -3.50, R) >droSec1.super_8 1735244 101 - 3762037 -CGCGAAGUUGACAGC--UGC-----AGUGCACCGGCAUUGAUGAUAACUAAUCCAAUG------AUG-AGAACUAA---GCCAUUGGAGAU-GUCCUCACCAAUGCCACGGCACUGCAU -.((..........))--(((-----(((((...(((((((.(((..((...(((((((------...-........---..)))))))...-))..))).)))))))...)))))))). ( -39.52, z-score = -4.84, R) >droSim1.chrX 15018510 103 - 17042790 -CGCGAAGUUGACAGCAGUGC-----AGUGCACCGGCAUUGAUGAUAACUAAUCCAAUG------AUG-AGAACCAA---UCCAUUGGAGAU-GUCCUCACCAAUGCCACGACACUGCAU -.((..........)).((((-----((((....(((((((.(((..((...(((((((------((.-.......)---).)))))))...-))..))).)))))))....)))))))) ( -37.00, z-score = -4.47, R) >droWil1.scaffold_181096 7956783 116 - 12416693 -CGCACAAUAUACGAUAUUGCUCACAAACACUUCGCCAUCGAUGAGAACUAAUCCAAUGUGUUCAAUGUUGGGCUCCAAUAACAUUGGAGCUGGAACUCACCAAUAUCGAGGUGUUG--- -.(..((((((...))))))..)...(((((((((..((.(.((((..(((..(((((((.....)))))))((((((((...)))))))))))..)))).).))..))))))))).--- ( -34.00, z-score = -3.13, R) >droVir3.scaffold_13042 4285165 88 + 5191987 ---------UGCUG---GUAC-----AGCGCAUCGGCAUCGAUGAGAACUAAUCCAAUG------UGA-UAUGCU-------CAUUGGAGAU-GCUAUCAUCAAUCGCGAGGGGCUGCAC ---------.....---((.(-----(((.(.((((.((.(((((.......(((((((------.(.-....).-------)))))))...-....))))).))).))).).)))).)) ( -26.04, z-score = -0.46, R) >droMoj3.scaffold_6328 1552363 88 + 4453435 ---------UGCUG---AUAC-----AGCGCAUCGACAUCGAUGAGAACUAAUCCAAUG------CGA-AAUACU-------CAUUGGAGAU-GCUUUCAUCAAUCGCGAGGGGCUGCAC ---------.....---...(-----(((.(.(((.....(((((((.(...(((((((------.(.-....).-------)))))))...-).))))))).....))).).))))... ( -27.80, z-score = -2.46, R) >consensus _CGCGAAGUUGAUAG__GUGC_____AGUGCACCGGCAUUGAUGAGAACUAAUCCAAUG______AUG_AGGGCU_C____CCAUUGGAGAU_GCCCUCACCAAUGUCGAGGCACUGCAU ..........................(((((..((.......(((.......(((((((.......................)))))))........))).......))..))))).... ( -9.44 = -9.89 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:26 2011