| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,452,781 – 19,452,883 |
| Length | 102 |
| Max. P | 0.999454 |

| Location | 19,452,781 – 19,452,883 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 70.56 |
| Shannon entropy | 0.59020 |
| G+C content | 0.40516 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.48 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
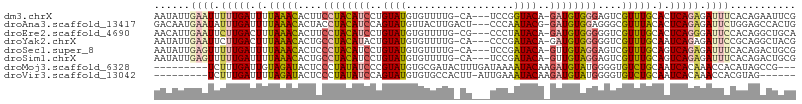
>dm3.chrX 19452781 102 + 22422827 AAUAUUGAAUUUUUGAUUUUAAACACUUCCUACAUCCUGUAUGUGUUUUG-CA---UCCGGUACA-GAUGUGGGAGUCGUUUGCACUCAGAGAUUUCACAGAAUUCG ......((((((((((.(.((((((((.((((((((.((((((((.....-))---)...)))))-))))))))))).))))).).))))))))))........... ( -29.60, z-score = -3.39, R) >droAna3.scaffold_13417 4492532 103 - 6960332 GACAAUGAAUAUUUGAUUUUAAACACUACCUACAUCCAGUAUGUUACUUGACU---CCCAAUACG-GAUGUGGAGGGCGUUUACACUCAGAGAUUCUGGAGCCACUG ......((((.(((((.(.(((((.((..((((((((.((((...........---....)))))-)))))))..)).))))).).))))).))))........... ( -25.76, z-score = -2.10, R) >droEre2.scaffold_4690 9688766 102 + 18748788 AACAUUGAAUUCUUGACUUUAAACACUUCCUACAUCCUGUAUGUGUUUUG-CG---CCCUAUACA-GAUGUGGGGGUCGUUUGCACUCAGGGAUUCCACAGGCUGCA ......((((((((((.(.((((((((.((((((((.((((((((.....-))---)...)))))-))))))))))).))))).).))))))))))........... ( -33.10, z-score = -2.79, R) >droYak2.chrX 9884799 102 - 21770863 AAUAUUGAAUUCUUGACUUUAAACACUGCCUACAUACUGUAUGUGUUUUG-CA---CCCGAUACA-GAUGUGGGGGUCGUUUGCAAUCAGAGAUUCCGCAGGCUACG ......((((..((((.(.((((((((.(((((((.(((((((((.....-))---)...)))))-))))))))))).))))).).))))..))))........... ( -28.80, z-score = -2.06, R) >droSec1.super_8 1735119 102 + 3762037 AAUAUUGAGUUUUUGAUUUUAAACACUCCCUACAUCCUGUAUGUGUUUUG-CA---UCCGAUACA-GUUGUAGGAGUCGUUUGCAGUCAGAGAUUUCACAGACUGCG ......((((((((((((.((((((((((.((((..(((((((((.....-))---)...)))))-).))))))))).))))).))))))))))))........... ( -31.10, z-score = -4.01, R) >droSim1.chrX 15018379 102 + 17042790 AAUAUUGAGUUUUUGAUUUUAAACACUGCCUACAUCCUGUAUGUGUUUUG-CA---UCCGAUACA-GUUGUAGGAGUCGUUUGCAGUCAGAGAUUUCACAGACUGCG ......((((((((((((.((((((((.((((((..(((((((((.....-))---)...)))))-).))))))))).))))).))))))))))))........... ( -31.10, z-score = -3.49, R) >droMoj3.scaffold_6328 1552211 95 - 4453435 ---------UCUUUGAUUGUAGAUACUCCCUAUAUCCCGUAUGUGCGAUACUUUGAUAAAAUACAAGAUGUAUGGGGUGUCUGCAAUCACAAACCACAUAGCCG--- ---------....((((((((((((((((.((((((..((((...(((....))).....))))..)))))).))))))))))))))))...............--- ( -33.90, z-score = -5.61, R) >droVir3.scaffold_13042 4284984 91 - 5191987 ---------UCUUUGAUUUUAGAUACUCCCUAUAUCCAGUAUGUGUGCCACUU-AUUGAAAUACAAGAUGUAUGGGGUGUCUGCAAUCACAAACCACGUAG------ ---------....(((((.((((((((((.(((((((((((.(((...))).)-))))........)))))).)))))))))).)))))............------ ( -26.30, z-score = -3.17, R) >consensus AAUAUUGAAUUUUUGAUUUUAAACACUCCCUACAUCCUGUAUGUGUUUUG_CA___UCCGAUACA_GAUGUAGGGGUCGUUUGCAAUCAGAGAUUCCACAGACUGCG ......((((.(((((.(.(((((....((((((((..((((..................))))..))))))))....))))).).))))).))))........... (-13.28 = -13.48 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:24 2011