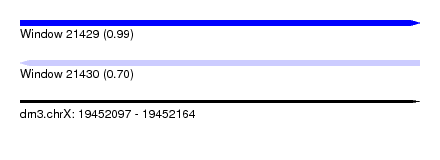
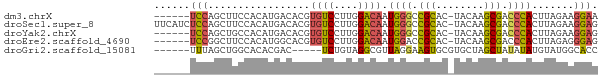
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,452,097 – 19,452,164 |
| Length | 67 |
| Max. P | 0.991093 |

| Location | 19,452,097 – 19,452,164 |
|---|---|
| Length | 67 |
| Sequences | 5 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 72.64 |
| Shannon entropy | 0.46427 |
| G+C content | 0.54258 |
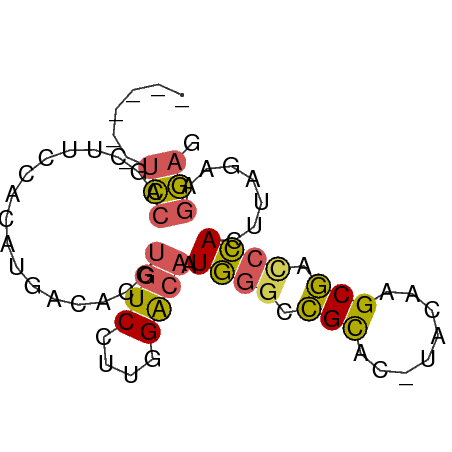
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -15.58 |
| Energy contribution | -17.14 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
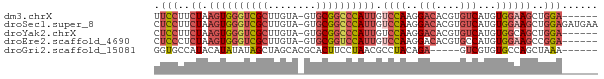
>dm3.chrX 19452097 67 + 22422827 UUCCUUCUAAGUGGGUCGCUUGUA-GUGCGGCCCAUUGUCCAAGGACACGUGUCAUGUGGAAGCUGGA------ .(((..((.((((((((((.....-..)))))))))).((((..(((....)))...))))))..)))------ ( -28.50, z-score = -3.33, R) >droSec1.super_8 1734328 73 + 3762037 CUCCUUCUAAGUGGGUCGCUUGUA-GUGCGGCCCAUUGUCCAAGGACACGUGUCAUGUGGAAGCUGGAGAUGAA ((((..((.((((((((((.....-..)))))))))).((((..(((....)))...))))))..))))..... ( -32.00, z-score = -3.92, R) >droYak2.chrX 9884359 67 - 21770863 CUCCUUCUAAGUGGGUCGCUUGUA-GUGCGGCCCAUUGUCCAAGGACACGUGUCAUGUGGCAGCUGGA------ .(((..((.((((((((((.....-..))))))))))(.(((..(((....)))...))))))..)))------ ( -26.80, z-score = -2.19, R) >droEre2.scaffold_4690 9688422 67 + 18748788 CUCCCUCUAAGUGGGUCGCUUGUA-GUGCGGUCCAUUGUCCAAGGACACGUGCCAUGUGGAAGCCGGA------ .(((..((.(((((..(((.....-..)))..))))).((((.((.(....)))...))))))..)))------ ( -20.90, z-score = -0.25, R) >droGri2.scaffold_15081 2229333 63 - 4274704 GGUGCCAUACAUAUAUAGCUAGCACGCACUUCCUAACGCCUACAGA-----GUCGUGUGCCAGCUAAA------ ...............(((((.((((((((((..((.....))..))-----)).)))))).)))))..------ ( -18.10, z-score = -1.80, R) >consensus CUCCUUCUAAGUGGGUCGCUUGUA_GUGCGGCCCAUUGUCCAAGGACACGUGUCAUGUGGAAGCUGGA______ .(((..((.((((((((((........)))))))))).((((..(((....)))...))))))..)))...... (-15.58 = -17.14 + 1.56)
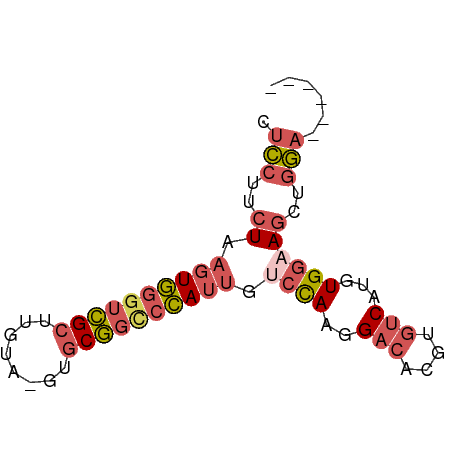
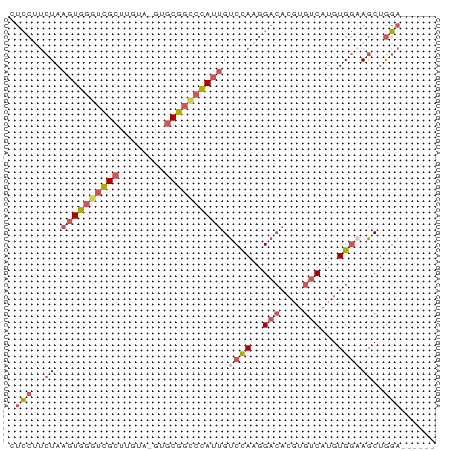
| Location | 19,452,097 – 19,452,164 |
|---|---|
| Length | 67 |
| Sequences | 5 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 72.64 |
| Shannon entropy | 0.46427 |
| G+C content | 0.54258 |
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -12.10 |
| Energy contribution | -11.90 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.702556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19452097 67 - 22422827 ------UCCAGCUUCCACAUGACACGUGUCCUUGGACAAUGGGCCGCAC-UACAAGCGACCCACUUAGAAGGAA ------(((....((((...(((....)))..))))...((((.(((..-.....))).)))).......))). ( -19.10, z-score = -1.81, R) >droSec1.super_8 1734328 73 - 3762037 UUCAUCUCCAGCUUCCACAUGACACGUGUCCUUGGACAAUGGGCCGCAC-UACAAGCGACCCACUUAGAAGGAG .....((((....((((...(((....)))..))))...((((.(((..-.....))).)))).......)))) ( -21.00, z-score = -2.20, R) >droYak2.chrX 9884359 67 + 21770863 ------UCCAGCUGCCACAUGACACGUGUCCUUGGACAAUGGGCCGCAC-UACAAGCGACCCACUUAGAAGGAG ------(((..((((((...(((....)))..)))....((((.(((..-.....))).))))..)))..))). ( -18.80, z-score = -1.24, R) >droEre2.scaffold_4690 9688422 67 - 18748788 ------UCCGGCUUCCACAUGGCACGUGUCCUUGGACAAUGGACCGCAC-UACAAGCGACCCACUUAGAGGGAG ------((((...((((...(((....)))..))))...)))).(((..-.....))).(((.......))).. ( -16.40, z-score = 0.20, R) >droGri2.scaffold_15081 2229333 63 + 4274704 ------UUUAGCUGGCACACGAC-----UCUGUAGGCGUUAGGAAGUGCGUGCUAGCUAUAUAUGUAUGGCACC ------..((((((((((.((..-----(((..........)))..)).))))))))))............... ( -19.20, z-score = -0.58, R) >consensus ______UCCAGCUUCCACAUGACACGUGUCCUUGGACAAUGGGCCGCAC_UACAAGCGACCCACUUAGAAGGAG ......(((.................((((....)))).((((.(((........))).)))).......))). (-12.10 = -11.90 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:23 2011