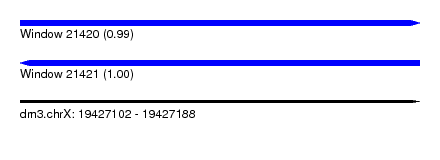
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,427,102 – 19,427,188 |
| Length | 86 |
| Max. P | 0.999939 |

| Location | 19,427,102 – 19,427,188 |
|---|---|
| Length | 86 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 64.36 |
| Shannon entropy | 0.74904 |
| G+C content | 0.46806 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -4.66 |
| Energy contribution | -4.67 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.17 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
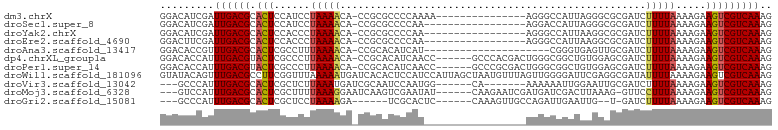
>dm3.chrX 19427102 86 + 22422827 GGACAUCGAUUGACGCACUCCAUCCUAAAACA-CCGCGCCCCAAAA---------------AGGGCCAUUAGGGCGCGAUCUUUUAAAAGAAGUCGUCAAAG .........((((((.(((.............-.(((((((.....---------------..........))))))).((((....))))))))))))).. ( -23.16, z-score = -2.06, R) >droSec1.super_8 1709170 84 + 3762037 GGACAUCGAUUGACGCACUCCAUCCUAAAACA-CCGCGCCCCAA-----------------AGGACCAUUAGGGCGCGAUCUUUUAAAAGAAGUCGUCAAAG .........((((((.(((.............-.(((((((...-----------------..........))))))).((((....))))))))))))).. ( -23.32, z-score = -2.84, R) >droYak2.chrX 9860989 84 - 21770863 GGACAUCGAUUGACGCACUCCACCCUAAAACA-CCGCGCCCCAA-----------------AGGGCCAUUAAGGCGCGAUCUUUUAAAAGAAGUCGUCAAAG .........((((((.(((.............-.((((((((..-----------------..)).......)))))).((((....))))))))))))).. ( -20.31, z-score = -1.67, R) >droEre2.scaffold_4690 9664288 84 + 18748788 GGACUUCGAUUGACGCACUCCACCCUAAAACA-CCGCGCCCCAA-----------------AGGGCCAUUAAGGCGCGAUCUUUUAAAAGAAGUCGUCAAAG .........((((((.(((.............-.((((((((..-----------------..)).......)))))).((((....))))))))))))).. ( -20.31, z-score = -1.45, R) >droAna3.scaffold_13417 4455282 80 - 6960332 GGACACCGUUUGACGCACUCGCCUUUAAAACA-CCGCACAUCAU---------------------CGGGUGAGUUGCGAUCUUUUAAAAGAAGUCGUCAAAG ........((((((((((((((((........-...........---------------------.))))))).)))((.((((.....)))))))))))). ( -21.05, z-score = -1.69, R) >dp4.chrXL_group1a 3766328 95 - 9151740 GGACACCAUUUGACGUACUCGCCCUUAAAACA-CCGCACAUCAACC------GCCCACGACUGGGCGGCUGUGGAGCGAUCUUUUAAAAGAAGUCGUCAAAG ........(((((((.((((....((((((..-.(((.((.((.((------(((((....))))))).))))..)))...))))))..).)))))))))). ( -31.50, z-score = -3.30, R) >droPer1.super_14 134966 95 - 2168203 GGACACCAUUUGACGUACUCGCCCUUAAAACA-CCGCACAUCAACC------GCCCGCGACUGGGCGGCUGUGGAGCGAUCUUUUAAAAGAAGUCGUCAAAG ........(((((((.((((....((((((..-.(((.((.((.((------(((((....))))))).))))..)))...))))))..).)))))))))). ( -30.80, z-score = -2.58, R) >droWil1.scaffold_181096 7924110 102 + 12416693 GUAUACAGUUUGACGCCUUCGGUUUAAAAAUGAUCACACUCCAUCCAUUAGCUAAUGUUUAGUUGGGGAUUCGAGGCGAUAUUUUAAAAGAAGUCGUCAAAG ........(((((((.((((..(((((((...(((.(.(((.((((.(((((((.....)))))))))))..)))).))).))))))).)))).))))))). ( -31.00, z-score = -4.11, R) >droVir3.scaffold_13042 4243972 86 - 5191987 ---GCCCAUUUGACGCACUCGCUCUUAAAUGAUCGCAAUCCAAUGG------CA-------AAAAAAUUGGAAUUGCGAUCUUUUAAAAGAAGUCGUCAAAG ---.....(((((((.(((..((.(((((.((((((((((((((..------..-------.....))))).))))))))).))))).)).)))))))))). ( -32.10, z-score = -6.38, R) >droMoj3.scaffold_6328 1519866 92 - 4453435 ---GUCCAUUUGACGCACUCGCUUUUAAAGGAAUCAAGUCGAAUAU------CAAGAAUCGAUGAUCGACUUAAAG-GUUCCUUUAAAAGAAGUCGUCAAAG ---.....(((((((.(((..(((((((((((((((((((((.(((------(.......)))).)))))))...)-))))))))))))).)))))))))). ( -38.60, z-score = -7.94, R) >droGri2.scaffold_15081 2200176 84 - 4274704 ---GCCCAUUUGACGCACUCGCUCCUAAAAGA------UCGCACUC------CAAAGUUGCCAGAUUGAAUUG--U-GAUCUUUUAAAAGAAGUCGUCAAAG ---.....(((((((.(((..((..(((((((------(((((...------...((((....))))....))--)-)))))))))..)).)))))))))). ( -24.60, z-score = -4.30, R) >consensus GGACACCGUUUGACGCACUCGCUCUUAAAACA_CCGCACCCCAA_________________AGGGCCGUUAAGGCGCGAUCUUUUAAAAGAAGUCGUCAAAG .........((((((.((((.....................................................................).))))))))).. ( -4.66 = -4.67 + 0.01)
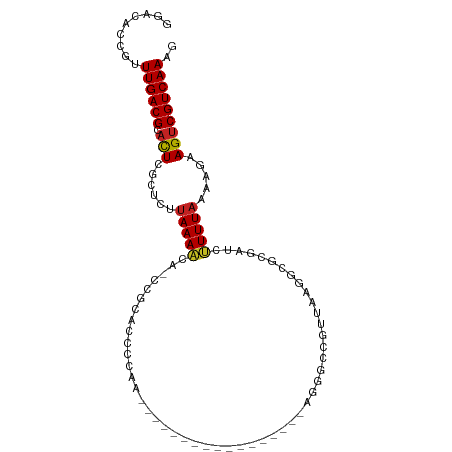
| Location | 19,427,102 – 19,427,188 |
|---|---|
| Length | 86 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 64.36 |
| Shannon entropy | 0.74904 |
| G+C content | 0.46806 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -8.64 |
| Energy contribution | -8.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -5.46 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.04 |
| SVM RNA-class probability | 0.999939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19427102 86 - 22422827 CUUUGACGACUUCUUUUAAAAGAUCGCGCCCUAAUGGCCCU---------------UUUUGGGGCGCGG-UGUUUUAGGAUGGAGUGCGUCAAUCGAUGUCC ..(((((((((((((((((((.((((((((((((.......---------------..)))))))))))-).)))))))).))))).))))))......... ( -40.90, z-score = -5.88, R) >droSec1.super_8 1709170 84 - 3762037 CUUUGACGACUUCUUUUAAAAGAUCGCGCCCUAAUGGUCCU-----------------UUGGGGCGCGG-UGUUUUAGGAUGGAGUGCGUCAAUCGAUGUCC ..(((((((((((((((((((.((((((((((((.......-----------------)))))))))))-).)))))))).))))).))))))......... ( -41.40, z-score = -6.45, R) >droYak2.chrX 9860989 84 + 21770863 CUUUGACGACUUCUUUUAAAAGAUCGCGCCUUAAUGGCCCU-----------------UUGGGGCGCGG-UGUUUUAGGGUGGAGUGCGUCAAUCGAUGUCC ..(((((((((((((((((((.((((((((((((.......-----------------)))))))))))-).)))))))).))))).))))))......... ( -37.80, z-score = -4.88, R) >droEre2.scaffold_4690 9664288 84 - 18748788 CUUUGACGACUUCUUUUAAAAGAUCGCGCCUUAAUGGCCCU-----------------UUGGGGCGCGG-UGUUUUAGGGUGGAGUGCGUCAAUCGAAGUCC ..(((((((((((((((((((.((((((((((((.......-----------------)))))))))))-).)))))))).))))).))))))......... ( -37.80, z-score = -4.70, R) >droAna3.scaffold_13417 4455282 80 + 6960332 CUUUGACGACUUCUUUUAAAAGAUCGCAACUCACCCG---------------------AUGAUGUGCGG-UGUUUUAAAGGCGAGUGCGUCAAACGGUGUCC .((((((((((((((((((((.((((((..(((....---------------------.)))..)))))-).))))))))).)))).)))))))........ ( -29.40, z-score = -3.65, R) >dp4.chrXL_group1a 3766328 95 + 9151740 CUUUGACGACUUCUUUUAAAAGAUCGCUCCACAGCCGCCCAGUCGUGGGC------GGUUGAUGUGCGG-UGUUUUAAGGGCGAGUACGUCAAAUGGUGUCC .((((((((((((((((((((.(((((..((((((((((((....)))))------))))).)).))))-).))))))))).)))).)))))))........ ( -46.60, z-score = -6.09, R) >droPer1.super_14 134966 95 + 2168203 CUUUGACGACUUCUUUUAAAAGAUCGCUCCACAGCCGCCCAGUCGCGGGC------GGUUGAUGUGCGG-UGUUUUAAGGGCGAGUACGUCAAAUGGUGUCC .((((((((((((((((((((.(((((..(((((((((((......))))------))))).)).))))-).))))))))).)))).)))))))........ ( -46.00, z-score = -5.59, R) >droWil1.scaffold_181096 7924110 102 - 12416693 CUUUGACGACUUCUUUUAAAAUAUCGCCUCGAAUCCCCAACUAAACAUUAGCUAAUGGAUGGAGUGUGAUCAUUUUUAAACCGAAGGCGUCAAACUGUAUAC .(((((((.((((.(((((((.((((((((..((((....(((.....))).....)))).))).)))))...)))))))..)))).)))))))........ ( -27.20, z-score = -3.91, R) >droVir3.scaffold_13042 4243972 86 + 5191987 CUUUGACGACUUCUUUUAAAAGAUCGCAAUUCCAAUUUUUU-------UG------CCAUUGGAUUGCGAUCAUUUAAGAGCGAGUGCGUCAAAUGGGC--- .(((((((((((((((((((.(((((((((.(((((.....-------..------..)))))))))))))).)))))))).)))).))))))).....--- ( -35.40, z-score = -6.08, R) >droMoj3.scaffold_6328 1519866 92 + 4453435 CUUUGACGACUUCUUUUAAAGGAAC-CUUUAAGUCGAUCAUCGAUUCUUG------AUAUUCGACUUGAUUCCUUUAAAAGCGAGUGCGUCAAAUGGAC--- .(((((((((((((((((((((((.-..(((((((((..(((((...)))------))..))))))))))))))))))))).)))).))))))).....--- ( -38.40, z-score = -7.40, R) >droGri2.scaffold_15081 2200176 84 + 4274704 CUUUGACGACUUCUUUUAAAAGAUC-A--CAAUUCAAUCUGGCAACUUUG------GAGUGCGA------UCUUUUAGGAGCGAGUGCGUCAAAUGGGC--- .((((((((((((((((((((((((-.--((.(((((...(....).)))------)).)).))------))))))))))).)))).))))))).....--- ( -33.40, z-score = -5.37, R) >consensus CUUUGACGACUUCUUUUAAAAGAUCGCGCCACAACGGCCCU_________________UUGGGGCGCGG_UGUUUUAAGAGCGAGUGCGUCAAACGGUGUCC ..((((((((((..(((((((.((((........................................))).).)))))))...)))).))))))......... ( -8.64 = -8.95 + 0.31)
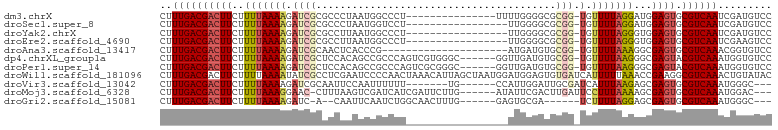

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:16 2011