
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,009,751 – 12,009,846 |
| Length | 95 |
| Max. P | 0.694519 |

| Location | 12,009,751 – 12,009,846 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 68.27 |
| Shannon entropy | 0.60964 |
| G+C content | 0.28238 |
| Mean single sequence MFE | -13.39 |
| Consensus MFE | -6.57 |
| Energy contribution | -6.54 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
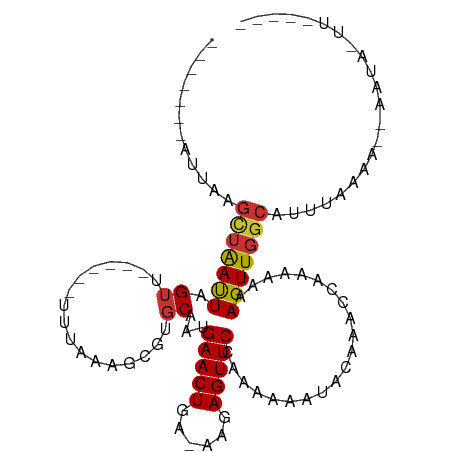
>dm3.chr2L 12009751 95 + 23011544 --ACGUAAUUAAGCUAAUUAGUU------GU-AGGUCGUGCAAUGAACUGA-AAGAGUUCCAAAAAAUACAAACCAAAAAAAGUUGGCAUUAAAAA--AUUAUUAAA--- --......(((((((((((.(((------((-(.....))))))(((((..-...))))).....................))))))).))))...--.........--- ( -13.80, z-score = -0.94, R) >droSim1.chr2L 11813889 97 + 22036055 AUACAUA-UUAAGCUAAUUAGUU------UUUAGGUCGUGCAAUGAACUGA-AAGAGUUCCAAAAAAUACAAACCAAAAAAAGUUGGCAUUCAAAA--AUUAAUCAA--- .......-....((((((...((------(((.(((.(((....(((((..-...))))).......)))..))).))))).))))))........--.........--- ( -13.70, z-score = -1.05, R) >droYak2.chr2L 8428543 83 + 22324452 -------AUUACGCUAAUUAGUU-------GUAAACAAUGCAAUGAACUGA-AAGAGUUCCAAAAAAUACAAACCAAAAAAAGUUGGCAUUUAAAA--AC---------- -------.....(((((((.(((-------(((.....))))))(((((..-...))))).....................)))))))........--..---------- ( -13.90, z-score = -2.27, R) >droEre2.scaffold_4929 13236835 95 - 26641161 -------AUUAAGCUAAUUAGUUAAUAGUUGUAAGGCGUGCAAUGAACUGA-AAGAGUUCCAAAAAAUACAAACCAAAAAAAGUUGGCAUUGAAAA--ACUAGUG----- -------.(((((((((((........((((((.....))))))(((((..-...))))).....................))))))).))))...--.......----- ( -14.00, z-score = -0.38, R) >droAna3.scaffold_12916 6722895 95 - 16180835 -----UAUACGAGUUGAUUAGUUU--------UAAG-GAGCAAUGAACUUG-AAGAGUUCCAAAAAAUAAACACCAAAAAAAGUUGGCAUUGAAAAGGAUUAUUUUUAAG -----....((((((.(((.((((--------....-))))))).))))))-(((((((((............((((......))))...(....))))..))))))... ( -14.10, z-score = -0.92, R) >dp4.chr4_group2 284670 101 - 1235136 -----AUUACGAGUUGAUUAGUUUAG---UUUUAAGUGCGCAACGAACUAC-AAAAGUUCAAAAAAAUACAAACCAAAAAAAGUUGGCAUUUGAAAAGAAUACCUUUAAA -----........((((.....((..---((((((((((.(((((((((..-...))))).......(........).....))))))))))))))..)).....)))). ( -16.10, z-score = -1.03, R) >droPer1.super_8 2945598 101 + 3966273 -----AUUACGAGUUGAUUAGUUUAG---UUCUAAGUGCGCAACGAACUAA-AAAAGUUCAAAAAAAUACAAACCAAAAAAAGUUGGCAUUUGAAAAGAAUACCUUUAAA -----...............((((..---(((.((((((.(((((((((..-...))))).......(........).....)))))))))))))..))))......... ( -14.60, z-score = -0.64, R) >droMoj3.scaffold_6500 26588637 81 - 32352404 ---------------AACUAGGUAUA----UAGGAUCAAGCAAUGAACUACUAAGAGUUCCAAAAAUACAAAACCAAAAAAAGUUGGCAUUCAAAAAGAA---------- ---------------..(((......----)))..((.......(((((......))))).............((((......))))..........)).---------- ( -6.90, z-score = 0.29, R) >consensus _______AUUAAGCUAAUUAGUU______UUUAAAGCGUGCAAUGAACUGA_AAGAGUUCCAAAAAAUACAAACCAAAAAAAGUUGGCAUUUAAAA__AAUA_UU_____ ............(((((((.((.................))...(((((......))))).....................)))))))...................... ( -6.57 = -6.54 + -0.03)


| Location | 12,009,751 – 12,009,846 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 68.27 |
| Shannon entropy | 0.60964 |
| G+C content | 0.28238 |
| Mean single sequence MFE | -11.79 |
| Consensus MFE | -7.65 |
| Energy contribution | -7.65 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.38 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.694519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12009751 95 - 23011544 ---UUUAAUAAU--UUUUUAAUGCCAACUUUUUUUGGUUUGUAUUUUUUGGAACUCUU-UCAGUUCAUUGCACGACCU-AC------AACUAAUUAGCUUAAUUACGU-- ---.........--...((((.((...........((((((((.......(((((...-..)))))..)))).)))).-..------.........))))))......-- ( -9.40, z-score = 0.27, R) >droSim1.chr2L 11813889 97 - 22036055 ---UUGAUUAAU--UUUUGAAUGCCAACUUUUUUUGGUUUGUAUUUUUUGGAACUCUU-UCAGUUCAUUGCACGACCUAAA------AACUAAUUAGCUUAA-UAUGUAU ---(((((((.(--((((((((((.((((......)))).))))))....(((((...-..)))))............)))------)).))))))).....-....... ( -13.80, z-score = -0.52, R) >droYak2.chr2L 8428543 83 - 22324452 ----------GU--UUUUAAAUGCCAACUUUUUUUGGUUUGUAUUUUUUGGAACUCUU-UCAGUUCAUUGCAUUGUUUAC-------AACUAAUUAGCGUAAU------- ----------((--(((.((((((.((((......)))).))))))...)))))....-...(((.((((..(((....)-------)).)))).))).....------- ( -11.20, z-score = -0.45, R) >droEre2.scaffold_4929 13236835 95 + 26641161 -----CACUAGU--UUUUCAAUGCCAACUUUUUUUGGUUUGUAUUUUUUGGAACUCUU-UCAGUUCAUUGCACGCCUUACAACUAUUAACUAAUUAGCUUAAU------- -----..(((((--(....(((((.((((......)))).))))).....(((((...-..))))).........................))))))......------- ( -11.10, z-score = -0.16, R) >droAna3.scaffold_12916 6722895 95 + 16180835 CUUAAAAAUAAUCCUUUUCAAUGCCAACUUUUUUUGGUGUUUAUUUUUUGGAACUCUU-CAAGUUCAUUGCUC-CUUA--------AAACUAAUCAACUCGUAUA----- ...((((((((.........(((((((......)))))))))))))))..(((((...-..))))).......-....--------...................----- ( -9.50, z-score = -0.68, R) >dp4.chr4_group2 284670 101 + 1235136 UUUAAAGGUAUUCUUUUCAAAUGCCAACUUUUUUUGGUUUGUAUUUUUUUGAACUUUU-GUAGUUCGUUGCGCACUUAAAA---CUAAACUAAUCAACUCGUAAU----- ......((((((.......)))))).......((((((((.........((((((...-..))))))...........)))---)))))................----- ( -14.45, z-score = -0.64, R) >droPer1.super_8 2945598 101 - 3966273 UUUAAAGGUAUUCUUUUCAAAUGCCAACUUUUUUUGGUUUGUAUUUUUUUGAACUUUU-UUAGUUCGUUGCGCACUUAGAA---CUAAACUAAUCAACUCGUAAU----- ......((((((.......)))))).......((((((((.........((((((...-..))))))...........)))---)))))................----- ( -14.85, z-score = -0.76, R) >droMoj3.scaffold_6500 26588637 81 + 32352404 ----------UUCUUUUUGAAUGCCAACUUUUUUUGGUUUUGUAUUUUUGGAACUCUUAGUAGUUCAUUGCUUGAUCCUA----UAUACCUAGUU--------------- ----------............(((((......)))))...((((....(((..((..(((((....))))).)))))..----.))))......--------------- ( -10.00, z-score = -0.13, R) >consensus _____AA_UAAU__UUUUAAAUGCCAACUUUUUUUGGUUUGUAUUUUUUGGAACUCUU_UCAGUUCAUUGCACGCUUUAAA______AACUAAUUAGCUUAAA_______ ......................(((((......)))))............(((((......)))))............................................ ( -7.65 = -7.65 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:13 2011