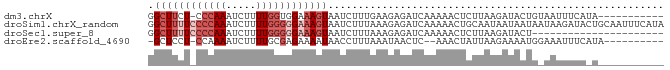
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,396,424 – 19,396,498 |
| Length | 74 |
| Max. P | 0.973885 |

| Location | 19,396,424 – 19,396,498 |
|---|---|
| Length | 74 |
| Sequences | 4 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 66.12 |
| Shannon entropy | 0.53882 |
| G+C content | 0.31204 |
| Mean single sequence MFE | -15.95 |
| Consensus MFE | -7.85 |
| Energy contribution | -8.60 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
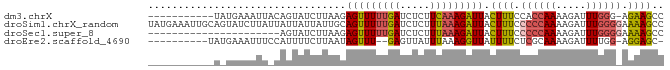
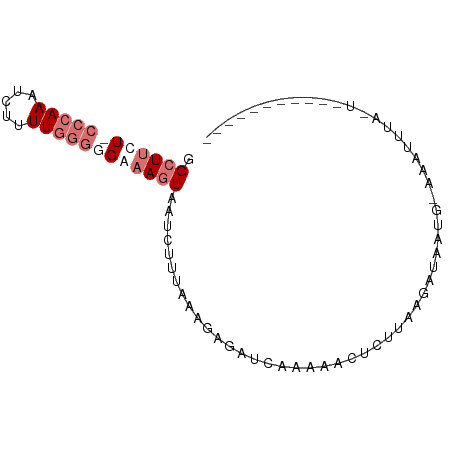
>dm3.chrX 19396424 74 + 22422827 -----------UAUGAAAUUACAGUAUCUUAAGAGUUUUUGAUCUCUUCAAAGAUUACUUUCCACCAAAAGAUUUGGG-AGAAGCC -----------............((.((((((((((((((((.....))))))))).)))....((((.....)))))-))).)). ( -13.50, z-score = -0.74, R) >droSim1.chrX_random 5048114 86 + 5698898 UAUGAAAUUGCAGUAUCUUAUUAUUAUUAUUGCAGUUUUUGAUCUCUUUAAAGAUUACUUUCCCCCAAAAGAUUUGGGGAAAAGCC ...((((((((((((............))))))))))))((((((......))))))((((.((((((.....)))))).)))).. ( -24.40, z-score = -4.04, R) >droSec1.super_8 1679285 64 + 3762037 ----------------------AGUAUCUUAAGAGUUUUUGAUCUCUUUAAAGAUUACUUUCCCCCAAAAGAUUUGGGGAAAAGCC ----------------------...((((((((((........)))))..)))))..((((.((((((.....)))))).)))).. ( -17.20, z-score = -2.10, R) >droEre2.scaffold_4690 9634583 72 + 18748788 ----------UAUGAAAUUUCCAUUUUCUUAAUAGUUU--GAGUUAUUUAAAGGUUAUUUUCUCGCAAAAGAUUUUGG-AGGAGC- ----------.((((..(((..((...(((((....))--)))..))..)))..))))((((((.((((....)))))-))))).- ( -8.70, z-score = 0.73, R) >consensus ___________A_UAAAUUU_CAGUAUCUUAAGAGUUUUUGAUCUCUUUAAAGAUUACUUUCCCCCAAAAGAUUUGGG_AAAAGCC .................................(((((((((.....))))))))).((((.((((((.....)))))).)))).. ( -7.85 = -8.60 + 0.75)
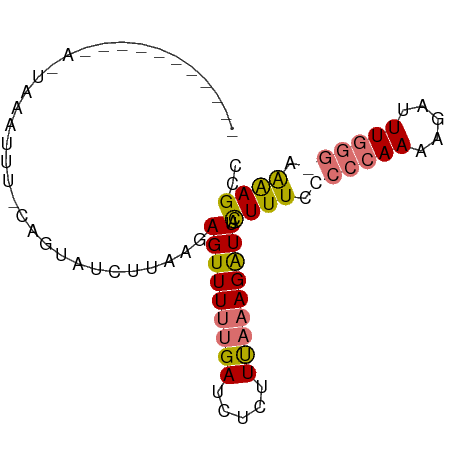
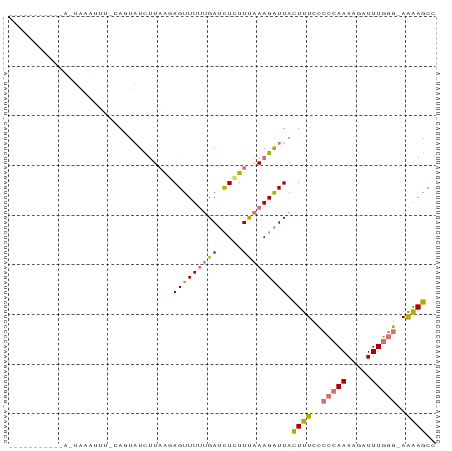
| Location | 19,396,424 – 19,396,498 |
|---|---|
| Length | 74 |
| Sequences | 4 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 66.12 |
| Shannon entropy | 0.53882 |
| G+C content | 0.31204 |
| Mean single sequence MFE | -16.02 |
| Consensus MFE | -7.18 |
| Energy contribution | -9.42 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19396424 74 - 22422827 GGCUUCU-CCCAAAUCUUUUGGUGGAAAGUAAUCUUUGAAGAGAUCAAAAACUCUUAAGAUACUGUAAUUUCAUA----------- ...(((.-.((((.....))))..)))((((.((((..(((((........)))))))))))))...........----------- ( -14.30, z-score = -1.07, R) >droSim1.chrX_random 5048114 86 - 5698898 GGCUUUUCCCCAAAUCUUUUGGGGGAAAGUAAUCUUUAAAGAGAUCAAAAACUGCAAUAAUAAUAAUAAGAUACUGCAAUUUCAUA .((((((((((((.....)))))))))))).(((((....))))).......((((..................))))........ ( -20.57, z-score = -2.90, R) >droSec1.super_8 1679285 64 - 3762037 GGCUUUUCCCCAAAUCUUUUGGGGGAAAGUAAUCUUUAAAGAGAUCAAAAACUCUUAAGAUACU---------------------- .((((((((((((.....)))))))))))).(((((..(((((........))))))))))...---------------------- ( -23.60, z-score = -4.59, R) >droEre2.scaffold_4690 9634583 72 - 18748788 -GCUCCU-CCAAAAUCUUUUGCGAGAAAAUAACCUUUAAAUAACUC--AAACUAUUAAGAAAAUGGAAAUUUCAUA---------- -....((-(((((....)))).))).....................--...(((((.....)))))..........---------- ( -5.60, z-score = -0.46, R) >consensus GGCUUCU_CCCAAAUCUUUUGGGGGAAAGUAAUCUUUAAAGAGAUCAAAAACUCUUAAGAUAAUG_AAAUUUA_U___________ .((((((((((((.....))))))))))))........................................................ ( -7.18 = -9.42 + 2.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:12 2011