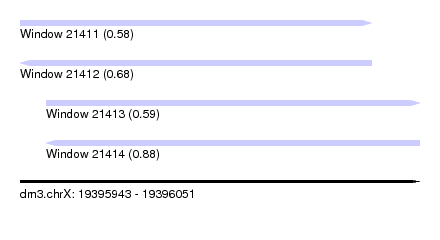
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,395,943 – 19,396,051 |
| Length | 108 |
| Max. P | 0.878162 |

| Location | 19,395,943 – 19,396,038 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 71.60 |
| Shannon entropy | 0.40872 |
| G+C content | 0.32969 |
| Mean single sequence MFE | -12.60 |
| Consensus MFE | -7.93 |
| Energy contribution | -7.55 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.583041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19395943 95 + 22422827 GAUAGUCGAAAAACAUGAACAAAUUACGAAUUUUACAUCCUCUGUUCCUCAACAAGGAACUACUAUGUACAUAUACAUACAUAUAUGUUCGUUAU ..............(((((((......................((((((.....))))))...((((((........))))))..)))))))... ( -15.70, z-score = -1.70, R) >droYak2.chrX 9830486 74 - 21770863 UACAGUCAUAAAAUAUGAACUAAUGACGAAUCUUAGAUUCCCCGUUCCCAAACAGGGAACUAC-AUGUUAGUCUU-------------------- .....(((((...)))))((((((...(((((...)))))...((((((.....))))))...-..))))))...-------------------- ( -16.50, z-score = -2.61, R) >droSec1.super_8 1678893 74 + 3762037 GAUAGAUAUAGAACAUGAACAAAUGACGAAUUUUACAUCCCCCGUUCCUCAACAAGGAACUAA-AUGUUAGUUUU-------------------- (((.(((((....(((......)))..................((((((.....))))))...-))))).)))..-------------------- ( -9.10, z-score = -0.57, R) >droSim1.chrX_random 5047668 74 + 5698898 GAUAGAUAUAGAACAUGAACAAAUGACGAAUUUUACAUACUCCGUUCCUCAACAAGGAACUAA-AUGUUAGUUUU-------------------- (((.(((((....(((......)))..................((((((.....))))))...-))))).)))..-------------------- ( -9.10, z-score = -0.37, R) >consensus GAUAGACAUAAAACAUGAACAAAUGACGAAUUUUACAUCCCCCGUUCCUCAACAAGGAACUAA_AUGUUAGUUUU____________________ ...........................................((((((.....))))))................................... ( -7.93 = -7.55 + -0.38)

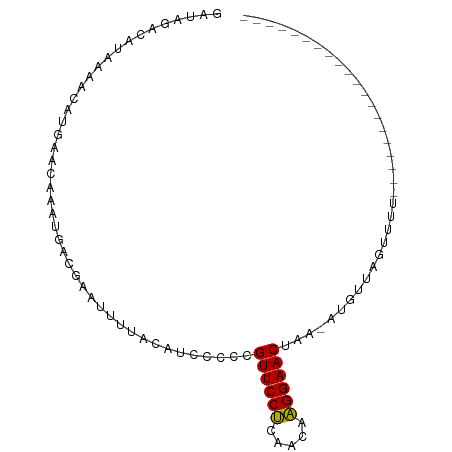

| Location | 19,395,943 – 19,396,038 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 71.60 |
| Shannon entropy | 0.40872 |
| G+C content | 0.32969 |
| Mean single sequence MFE | -16.91 |
| Consensus MFE | -10.65 |
| Energy contribution | -9.90 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19395943 95 - 22422827 AUAACGAACAUAUAUGUAUGUAUAUGUACAUAGUAGUUCCUUGUUGAGGAACAGAGGAUGUAAAAUUCGUAAUUUGUUCAUGUUUUUCGACUAUC .....((((...(((((((......)))))))...))))...(((((((((((..(((((..............))))).))))))))))).... ( -22.14, z-score = -1.82, R) >droYak2.chrX 9830486 74 + 21770863 --------------------AAGACUAACAU-GUAGUUCCCUGUUUGGGAACGGGGAAUCUAAGAUUCGUCAUUAGUUCAUAUUUUAUGACUGUA --------------------..((((((...-...((((((.....))))))((.(((((...))))).)).))))))................. ( -19.40, z-score = -1.88, R) >droSec1.super_8 1678893 74 - 3762037 --------------------AAAACUAACAU-UUAGUUCCUUGUUGAGGAACGGGGGAUGUAAAAUUCGUCAUUUGUUCAUGUUCUAUAUCUAUC --------------------......(((((-...(((((((...)))))))((.((((.....)))).))........)))))........... ( -13.50, z-score = -0.85, R) >droSim1.chrX_random 5047668 74 - 5698898 --------------------AAAACUAACAU-UUAGUUCCUUGUUGAGGAACGGAGUAUGUAAAAUUCGUCAUUUGUUCAUGUUCUAUAUCUAUC --------------------...........-...(((((((...)))))))((((((((..((((.....))))...))))))))......... ( -12.60, z-score = -1.13, R) >consensus ____________________AAAACUAACAU_GUAGUUCCUUGUUGAGGAACGGAGGAUGUAAAAUUCGUCAUUUGUUCAUGUUCUAUAACUAUC ...................................((((((.....))))))((((.(((..((((.....))))...))).))))......... (-10.65 = -9.90 + -0.75)

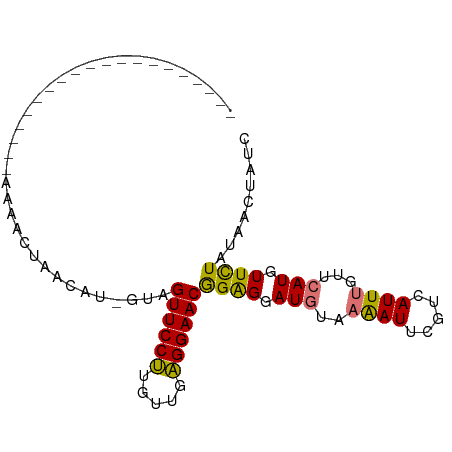
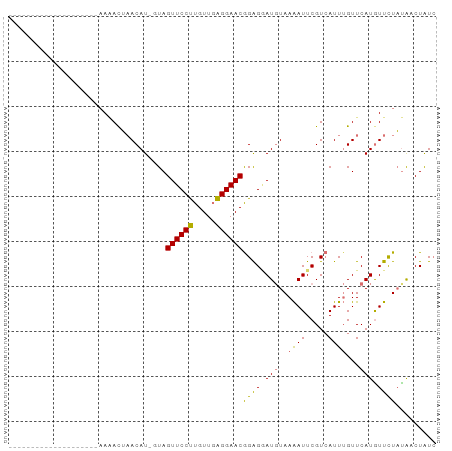
| Location | 19,395,950 – 19,396,051 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 70.92 |
| Shannon entropy | 0.37688 |
| G+C content | 0.33148 |
| Mean single sequence MFE | -14.30 |
| Consensus MFE | -9.01 |
| Energy contribution | -8.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.592117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19395950 101 + 22422827 GAAAAACAUGAACAAAUUACGAAUUUUACAUCCUCUGUUCCUCAACAAGGAACUACUAUGUACAUAUACAUACAUAUAUGUUCGUUAUGACACUUACUGAU ......((((........(((((...(((((.....((((((.....))))))....)))))((((((......)))))))))))))))............ ( -15.90, z-score = -1.90, R) >droYak2.chrX 9830493 80 - 21770863 AUAAAAUAUGAACUAAUGACGAAUCUUAGAUUCCCCGUUCCCAAACAGGGAACU---------------------ACAUGUUAGUCUUGACACUUUCUGAU ....((((((..........(((((...)))))...((((((.....)))))).---------------------.))))))................... ( -15.70, z-score = -1.75, R) >droSim1.chrX_random 5047675 80 + 5698898 AUAGAACAUGAACAAAUGACGAAUUUUACAUACUCCGUUCCUCAACAAGGAACU---------------------AAAUGUUAGUUUUGACACUUACUGAU .(((..(((......))).((((....((((.....((((((.....)))))).---------------------..))))....)))).......))).. ( -11.30, z-score = -0.78, R) >consensus AUAAAACAUGAACAAAUGACGAAUUUUACAUACUCCGUUCCUCAACAAGGAACU_____________________AAAUGUUAGUUUUGACACUUACUGAU ....................................((((((.....)))))).........................((((......))))......... ( -9.01 = -8.57 + -0.44)

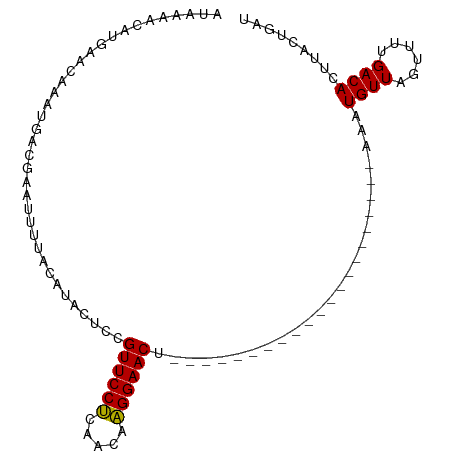
| Location | 19,395,950 – 19,396,051 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 70.92 |
| Shannon entropy | 0.37688 |
| G+C content | 0.33148 |
| Mean single sequence MFE | -20.13 |
| Consensus MFE | -11.39 |
| Energy contribution | -11.07 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19395950 101 - 22422827 AUCAGUAAGUGUCAUAACGAACAUAUAUGUAUGUAUAUGUACAUAGUAGUUCCUUGUUGAGGAACAGAGGAUGUAAAAUUCGUAAUUUGUUCAUGUUUUUC .((((((((.((....))((((...(((((((......)))))))...)))))))))))).(((((((..(((.......)))..)))))))......... ( -22.00, z-score = -1.67, R) >droYak2.chrX 9830493 80 + 21770863 AUCAGAAAGUGUCAAGACUAACAUGU---------------------AGUUCCCUGUUUGGGAACGGGGAAUCUAAGAUUCGUCAUUAGUUCAUAUUUUAU .....(((((((...((((((.....---------------------.((((((.....))))))((.(((((...))))).)).)))))).))))))).. ( -22.20, z-score = -2.81, R) >droSim1.chrX_random 5047675 80 - 5698898 AUCAGUAAGUGUCAAAACUAACAUUU---------------------AGUUCCUUGUUGAGGAACGGAGUAUGUAAAAUUCGUCAUUUGUUCAUGUUCUAU .....(((((((........))))))---------------------)(((((((...)))))))((((((((..((((.....))))...)))))))).. ( -16.20, z-score = -1.87, R) >consensus AUCAGUAAGUGUCAAAACUAACAUAU_____________________AGUUCCUUGUUGAGGAACGGAGAAUGUAAAAUUCGUCAUUUGUUCAUGUUUUAU ........(((....((((((...........................((((((.....))))))...((((.....))))....)))))))))....... (-11.39 = -11.07 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:11 2011