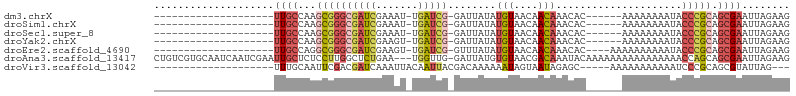
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,389,567 – 19,389,645 |
| Length | 78 |
| Max. P | 0.976429 |

| Location | 19,389,567 – 19,389,645 |
|---|---|
| Length | 78 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.19 |
| Shannon entropy | 0.45284 |
| G+C content | 0.38504 |
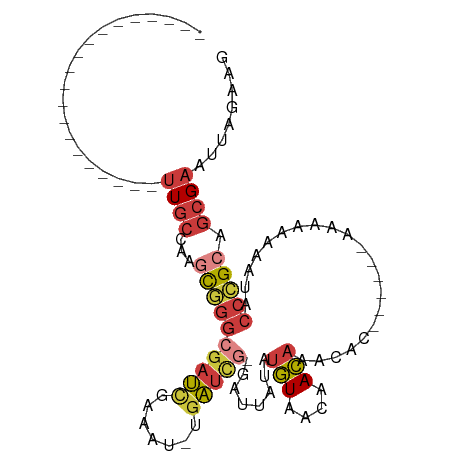
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -9.78 |
| Energy contribution | -10.69 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
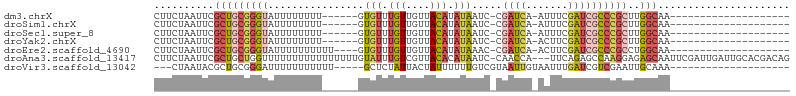
>dm3.chrX 19389567 78 + 22422827 CUUCUAAUUCGCUGCGGGUAUUUUUUUU------GUGUUUGUUGUUACAUAUAAUC-CGAUCA-AUUUCGAUCGCCCGCUUGGCAA-------------------- ..........(((((((((........(------((((........))))).....-.((((.-.....))))))))))..)))..-------------------- ( -21.20, z-score = -3.19, R) >droSim1.chrX 14980738 78 + 17042790 CUUCUAAUUCGCUGCGGGUAUUUUUUUU------GUGUUUGUUGUUACAUAUAAUC-CGAUCA-AUUUCGAUCGCCCGCUUGGCAA-------------------- ..........(((((((((........(------((((........))))).....-.((((.-.....))))))))))..)))..-------------------- ( -21.20, z-score = -3.19, R) >droSec1.super_8 1672567 78 + 3762037 CUUCUAAUUCGCUGCGGGUAUUUUUUUU------GUGUUUGUUGUUACAUAUAAUC-CGAUCA-AUUUCGAUCGCCCGCUUGGCAA-------------------- ..........(((((((((........(------((((........))))).....-.((((.-.....))))))))))..)))..-------------------- ( -21.20, z-score = -3.19, R) >droYak2.chrX 9824435 78 - 21770863 CUUCUAAUUCGCUGCGGGUAUUUUUUUU------GUGUUUGUUGUUACAUAUAAUC-CGAUCA-ACUUCGAUCGCCCGCUUGGCAA-------------------- ..........(((((((((........(------((((........))))).....-.((((.-.....))))))))))..)))..-------------------- ( -21.20, z-score = -3.24, R) >droEre2.scaffold_4690 9628098 80 + 18748788 CUUCUAAUUCGCUGCGGGUAUUUUUUUUUU----GUGUUUGUUGUUACAUAUAAAC-CGAUCA-ACUUCGAUCGCCCGCCUGGCAA-------------------- ..........(((((((((...........----(.((((((........))))))-)((((.-.....))))))))))..)))..-------------------- ( -21.80, z-score = -3.29, R) >droAna3.scaffold_13417 4405154 102 - 6960332 CUUCUAAUUCGCUGCUGGUUUUUUUUUUUUUUUUGUAUUUGUCGUUACACAUAAUC-CAACCA---UUCAGAGCCAAGGAGAGCAAUUCGAUUGAUUGCACGACAG ..(((..((.(((.((((...............((((........)))).......-......---.))))))).))..)))((((((.....))))))....... ( -15.45, z-score = 0.40, R) >droVir3.scaffold_13042 4191679 78 - 5191987 ---CUAAUACGCUGCGGGAUUUUUUUUUUU-----GCUCUAUUACUAUUUUUUGUCGUAAUUGUAAUUUGAUCGUCGAAUUGCAAA-------------------- ---....((((..((((((.......))))-----))......((........)))))).((((((((((.....)))))))))).-------------------- ( -13.50, z-score = -1.52, R) >consensus CUUCUAAUUCGCUGCGGGUAUUUUUUUU______GUGUUUGUUGUUACAUAUAAUC_CGAUCA_AUUUCGAUCGCCCGCUUGGCAA____________________ ..........(((((((((................(((.(((....))).))).....((((.......))))))))))..)))...................... ( -9.78 = -10.69 + 0.90)
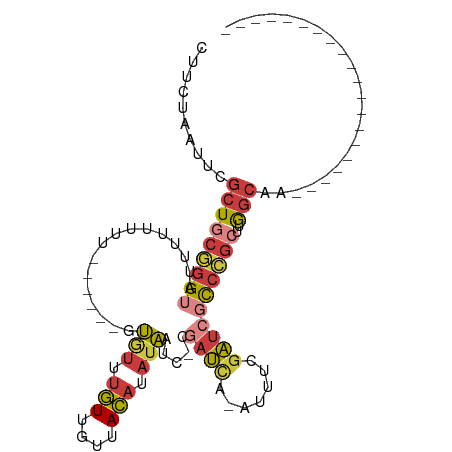
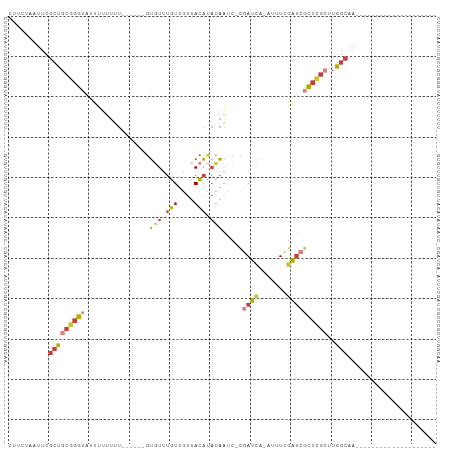
| Location | 19,389,567 – 19,389,645 |
|---|---|
| Length | 78 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.19 |
| Shannon entropy | 0.45284 |
| G+C content | 0.38504 |
| Mean single sequence MFE | -16.14 |
| Consensus MFE | -8.06 |
| Energy contribution | -8.90 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19389567 78 - 22422827 --------------------UUGCCAAGCGGGCGAUCGAAAU-UGAUCG-GAUUAUAUGUAACAACAAACAC------AAAAAAAAUACCCGCAGCGAAUUAGAAG --------------------((((...(((((((((((....-))))))-.......(((....))).....------..........))))).))))........ ( -18.10, z-score = -3.09, R) >droSim1.chrX 14980738 78 - 17042790 --------------------UUGCCAAGCGGGCGAUCGAAAU-UGAUCG-GAUUAUAUGUAACAACAAACAC------AAAAAAAAUACCCGCAGCGAAUUAGAAG --------------------((((...(((((((((((....-))))))-.......(((....))).....------..........))))).))))........ ( -18.10, z-score = -3.09, R) >droSec1.super_8 1672567 78 - 3762037 --------------------UUGCCAAGCGGGCGAUCGAAAU-UGAUCG-GAUUAUAUGUAACAACAAACAC------AAAAAAAAUACCCGCAGCGAAUUAGAAG --------------------((((...(((((((((((....-))))))-.......(((....))).....------..........))))).))))........ ( -18.10, z-score = -3.09, R) >droYak2.chrX 9824435 78 + 21770863 --------------------UUGCCAAGCGGGCGAUCGAAGU-UGAUCG-GAUUAUAUGUAACAACAAACAC------AAAAAAAAUACCCGCAGCGAAUUAGAAG --------------------((((...(((((((((((....-))))))-.......(((....))).....------..........))))).))))........ ( -18.10, z-score = -2.74, R) >droEre2.scaffold_4690 9628098 80 - 18748788 --------------------UUGCCAGGCGGGCGAUCGAAGU-UGAUCG-GUUUAUAUGUAACAACAAACAC----AAAAAAAAAAUACCCGCAGCGAAUUAGAAG --------------------((((...(((((((((((....-))))))-.......(((....))).....----............))))).))))........ ( -19.20, z-score = -2.48, R) >droAna3.scaffold_13417 4405154 102 + 6960332 CUGUCGUGCAAUCAAUCGAAUUGCUCUCCUUGGCUCUGAA---UGGUUG-GAUUAUGUGUAACGACAAAUACAAAAAAAAAAAAAAAACCAGCAGCGAAUUAGAAG .(((((((((...((((.((((..((...........)).---.)))).-))))...))).))))))....................................... ( -16.10, z-score = 0.02, R) >droVir3.scaffold_13042 4191679 78 + 5191987 --------------------UUUGCAAUUCGACGAUCAAAUUACAAUUACGACAAAAAAUAGUAAUAGAGC-----AAAAAAAAAAAUCCCGCAGCGUAUUAG--- --------------------(((((((((.(.....).))))...(((((...........)))))...))-----)))........................--- ( -5.30, z-score = 0.28, R) >consensus ____________________UUGCCAAGCGGGCGAUCGAAAU_UGAUCG_GAUUAUAUGUAACAACAAACAC______AAAAAAAAUACCCGCAGCGAAUUAGAAG ....................((((...(((((.(((((.....))))).........(((....))).....................))))).))))........ ( -8.06 = -8.90 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:06 2011