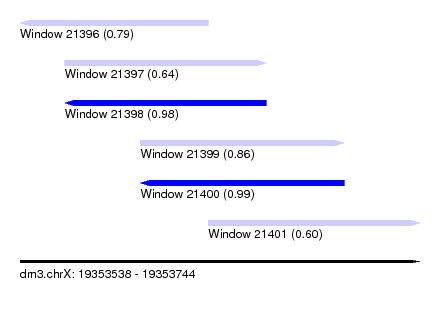
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,353,538 – 19,353,744 |
| Length | 206 |
| Max. P | 0.989256 |

| Location | 19,353,538 – 19,353,635 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 90.07 |
| Shannon entropy | 0.13245 |
| G+C content | 0.42465 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.794529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19353538 97 - 22422827 GCAAGAACACUUUCACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGC-AAACUUCUG------UGCACUCCUGCCAAAUAC ((((((((((............))))))))))...(((..(((..(((.((.(((((((........)))-)))).)).)------)).)))..)))....... ( -22.80, z-score = -1.74, R) >droYak2.chrX 9788906 94 + 21770863 GCAAGAACACUUC--UUCACAUGUGUUUUUGUUUCGC-AAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGC-AAACUUCUG------CGCACUCCUGCCAAAUAU ((((((((((...--.......))))))))))...((-((((((...((((((..........)))))).-.))))).))------)(((....)))....... ( -23.00, z-score = -2.34, R) >droEre2.scaffold_4690 9593527 101 - 18748788 GCAAGAACACUUC--UUCACAUGUGUUUUUGUUUCGC-AAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAAAACUUCUGCACUUCCGCACUCCUGCCAAAUAU ((((((((((...--.......))))))))))...((-((((((.(.((((((..........)))))).).))))).)))......(((....)))....... ( -23.30, z-score = -2.49, R) >consensus GCAAGAACACUUC__UUCACAUGUGUUUUUGUUUCGC_AAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGC_AAACUUCUG______CGCACUCCUGCCAAAUAU ((((((((((............)))))))))).......(((((...((((((..........))))))...)))))..........(((....)))....... (-18.30 = -18.30 + 0.00)



| Location | 19,353,561 – 19,353,665 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.96 |
| Shannon entropy | 0.15912 |
| G+C content | 0.45236 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -26.75 |
| Energy contribution | -26.95 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19353561 104 + 22422827 -GUUUGCUGGCGAAAGAGCAAACACCAAUAGAACUUUUGCGAAACAAAAACACAUGUGUGUG-----AAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCG---------- -(((((((..(....)))))))).......((((...(((((((((...((((....)))).-----....)))).)))))..)))).((((((....))))))......---------- ( -34.50, z-score = -2.59, R) >droSec1.super_8 1637266 109 + 3762037 -GUUUGCUGGCGAAAGAGCAAACACCAAUAGAACUUUUGCGAAACAAAAACACAUGUGUGUGUAAUGGAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCG---------- -(((((((..(....))))))))......(((((.(((.((..(((...(((....))).)))..)).))).))))).....((((..((((((....))))))..))))---------- ( -32.70, z-score = -1.55, R) >droSim1.chrX 14970608 109 + 17042790 -GUUUGCUGGCGAAAGAGCAAACACCAAUAGAACUUUUGCGAAACAAAAACACAUGUGUGUGUAAUGGAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCG---------- -(((((((..(....))))))))......(((((.(((.((..(((...(((....))).)))..)).))).))))).....((((..((((((....))))))..))))---------- ( -32.70, z-score = -1.55, R) >droYak2.chrX 9788929 101 - 21770863 -GUUUGCUGGCGAAAGAGCAAACACCAAUAGAACUUU-GCGAAACAAAAACACAUGUGAA-------GAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCG---------- -(((((((..(....)))))))).......((((..(-((((((((....((....))..-------....)))).)))))..)))).((((((....))))))......---------- ( -30.50, z-score = -1.94, R) >droEre2.scaffold_4690 9593556 112 + 18748788 GUUUUGCUGGCGAAAGAGCAAACACCAAUAGAACUUU-GCGAAACAAAAACACAUGUGAA-------GAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCGAGGAAAAGCG ((((((((..(....)))).....((..........(-((((((((....((....))..-------....)))).)))))(((((..((((((....))))))..))))))).))))). ( -29.90, z-score = -0.69, R) >consensus _GUUUGCUGGCGAAAGAGCAAACACCAAUAGAACUUUUGCGAAACAAAAACACAUGUGUGUG_____GAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCG__________ .(((((((..(....)))))))).................((..(((.(((((.................))))).)))..))(((..((((((....))))))..)))........... (-26.75 = -26.95 + 0.20)

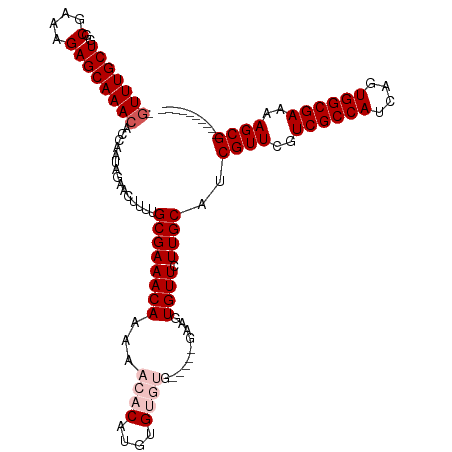
| Location | 19,353,561 – 19,353,665 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.96 |
| Shannon entropy | 0.15912 |
| G+C content | 0.45236 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -29.19 |
| Energy contribution | -29.19 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19353561 104 - 22422827 ----------CGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUU-----CACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAAAC- ----------.(((..((((((....))))))((((.((.((((((((((...-----.........)))))))))).))(((((...((....))..)))))...))))..)))....- ( -30.90, z-score = -2.23, R) >droSec1.super_8 1637266 109 - 3762037 ----------CGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUCCAUUACACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAAAC- ----------.(((..((((((....))))))((((.((.((((((((((.................)))))))))).))(((((...((....))..)))))...))))..)))....- ( -30.53, z-score = -2.17, R) >droSim1.chrX 14970608 109 - 17042790 ----------CGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUCCAUUACACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAAAC- ----------.(((..((((((....))))))((((.((.((((((((((.................)))))))))).))(((((...((....))..)))))...))))..)))....- ( -30.53, z-score = -2.17, R) >droYak2.chrX 9788929 101 + 21770863 ----------CGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUC-------UUCACAUGUGUUUUUGUUUCGC-AAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAAAC- ----------.(((..((((((....))))))((((.((.((((((((((...-------.......)))))))))).))((-(((..((....))..)))))...))))..)))....- ( -31.00, z-score = -2.51, R) >droEre2.scaffold_4690 9593556 112 - 18748788 CGCUUUUCCUCGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUC-------UUCACAUGUGUUUUUGUUUCGC-AAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAAAAC ((((.......(((((((((((....))))))....(((.((((((((((...-------.......)))))))))).))).-)))))......))))((((((........)))))).. ( -31.32, z-score = -2.13, R) >consensus __________CGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUC_____CACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAAAC_ ...........((.....))(((..((((.(((...(((.((((((((((.................)))))))))).))).....)))))))..)))((((((........)))))).. (-29.19 = -29.19 + -0.00)

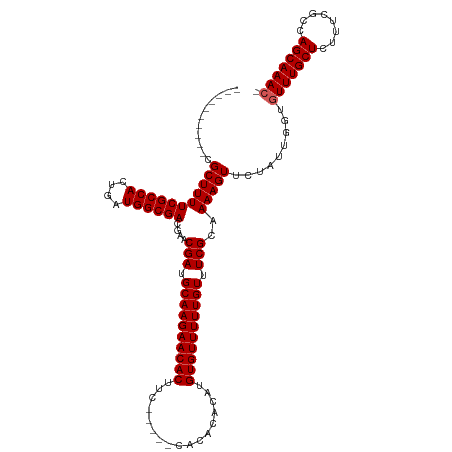

| Location | 19,353,600 – 19,353,705 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.25 |
| Shannon entropy | 0.17180 |
| G+C content | 0.38299 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -22.44 |
| Energy contribution | -22.84 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19353600 105 + 22422827 GAAACAAAAACACAUGUGUGUG-----AAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCGA----------CGAAAAGCUAAUAUGUGUAUAAAAAUGUUUUCCUUUGGU ((((((...((((((((.((..-----(........)..))(((((..((((((....))))))..)))))----------..........)))))))).......))))))........ ( -26.00, z-score = -1.68, R) >droSec1.super_8 1637305 108 + 3762037 GAAACAAAAACACAUGUGUGUGUAAUGGAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCGA----------CGAAAAGCUAAUAUGUGUAUAAAAAUGUUUUCCUUUG-- ((((((...((((((((..((....((.(((....))).))(((((..((((((....))))))..)))))----------......))..)))))))).......))))))......-- ( -27.80, z-score = -2.14, R) >droSim1.chrX 14970647 108 + 17042790 GAAACAAAAACACAUGUGUGUGUAAUGGAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCGA----------CGAAAAGCUAAUAUGUGUAUAAAAAUGUUUUCCUUUG-- ((((((...((((((((..((....((.(((....))).))(((((..((((((....))))))..)))))----------......))..)))))))).......))))))......-- ( -27.80, z-score = -2.14, R) >droYak2.chrX 9788967 102 - 21770863 GAAACAAAAACACAUGUGAA-------GAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCGA----------CGAAAAGCUAAUAUGUAUAUAAAAAUGUUUUUCUUUGU- ....(((.(((((.......-------...))))).)))..(((((..((((((....))))))..)))))----------.(((((((..................))))))).....- ( -24.87, z-score = -2.01, R) >droEre2.scaffold_4690 9593595 112 + 18748788 GAAACAAAAACACAUGUGAA-------GAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCGAGGAAAAGCGACGAAAAGCUAAUAUGUAUAUAAAAAUGUUUUCCUUUGU- ....(((.(((((.......-------...))))).)))..(((((..((((((....))))))..)))))((((((((........))).((((..........))))))))).....- ( -26.30, z-score = -1.53, R) >consensus GAAACAAAAACACAUGUGUGUG_____GAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCGA__________CGAAAAGCUAAUAUGUGUAUAAAAAUGUUUUCCUUUG__ ((((((...((((((((..............(((....)))(((((..((((((....))))))..)))))....................)))))))).......))))))........ (-22.44 = -22.84 + 0.40)



| Location | 19,353,600 – 19,353,705 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.25 |
| Shannon entropy | 0.17180 |
| G+C content | 0.38299 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -21.08 |
| Energy contribution | -20.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19353600 105 - 22422827 ACCAAAGGAAAACAUUUUUAUACACAUAUUAGCUUUUCG----------UCGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUU-----CACACACAUGUGUUUUUGUUUC ...................................((((----------(((((..(((....))).))))))))).((.((((((((((...-----.........)))))))))).)) ( -25.20, z-score = -3.00, R) >droSec1.super_8 1637305 108 - 3762037 --CAAAGGAAAACAUUUUUAUACACAUAUUAGCUUUUCG----------UCGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUCCAUUACACACACAUGUGUUUUUGUUUC --.................................((((----------(((((..(((....))).))))))))).((.((((((((((.................)))))))))).)) ( -24.83, z-score = -2.89, R) >droSim1.chrX 14970647 108 - 17042790 --CAAAGGAAAACAUUUUUAUACACAUAUUAGCUUUUCG----------UCGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUCCAUUACACACACAUGUGUUUUUGUUUC --.................................((((----------(((((..(((....))).))))))))).((.((((((((((.................)))))))))).)) ( -24.83, z-score = -2.89, R) >droYak2.chrX 9788967 102 + 21770863 -ACAAAGAAAAACAUUUUUAUAUACAUAUUAGCUUUUCG----------UCGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUC-------UUCACAUGUGUUUUUGUUUC -..................................((((----------(((((..(((....))).))))))))).((.((((((((((...-------.......)))))))))).)) ( -25.40, z-score = -3.24, R) >droEre2.scaffold_4690 9593595 112 - 18748788 -ACAAAGGAAAACAUUUUUAUAUACAUAUUAGCUUUUCGUCGCUUUUCCUCGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUC-------UUCACAUGUGUUUUUGUUUC -..................................(((((((((.......((.....)).......))))))))).((.((((((((((...-------.......)))))))))).)) ( -26.04, z-score = -2.68, R) >consensus __CAAAGGAAAACAUUUUUAUACACAUAUUAGCUUUUCG__________UCGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUC_____CACACACAUGUGUUUUUGUUUC ......(((((.(..................).)))))...........(((.((.((((((....)))))).)).))).((((((((((.................))))))))))... (-21.08 = -20.92 + -0.16)


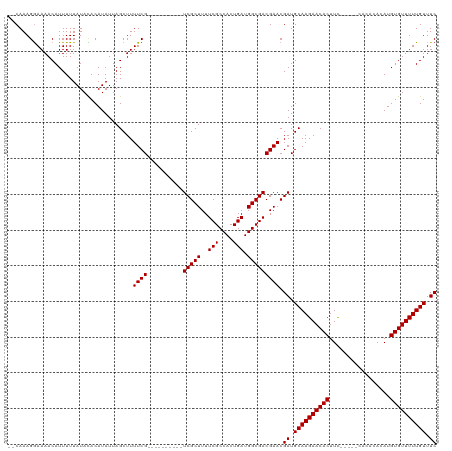
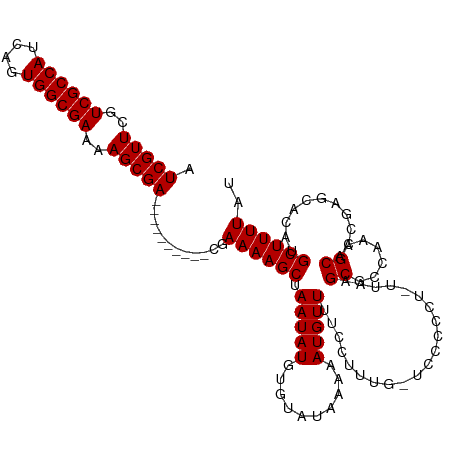
| Location | 19,353,635 – 19,353,744 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.38 |
| Shannon entropy | 0.17761 |
| G+C content | 0.43666 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.62 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19353635 109 + 22422827 AUCGUUCGUCGCCAUCAGUGGCGAAAAGCGA----------CGAAAAGCUAAUAUGUGUAUAAAAAUGUUUUCCUUUGGUCCCCCCUUUUAGCGGCAA-GCAACGAGCACAUGCUUUUAU .(((((..((((((....))))))..)))))----------..((((((....((((((.......(((((.((.((((.........)))).)).))-)))....)))))))))))).. ( -28.90, z-score = -1.73, R) >droSec1.super_8 1637345 104 + 3762037 AUCGUUCGUCGCCAUCAGUGGCGAAAAGCGA----------CGAAAAGCUAAUAUGUGUAUAAAAAUGUUUUCCUUUG-UCCCCUU-UUA-GCGCCAAAGCACCGAGC---UGCUUUUAU .(((((..((((((....))))))..)))))----------..((((((......((((.(((((..(..(......)-..)..))-)))-))))...(((.....))---))))))).. ( -24.70, z-score = -1.53, R) >droSim1.chrX 14970687 107 + 17042790 AUCGUUCGUCGCCAUCAGUGGCGAAAAGCGA----------CGAAAAGCUAAUAUGUGUAUAAAAAUGUUUUCCUUUG-UCCCCUU-UUA-GCGCCAAAGCACCGAGCACAUGCUUUUAU .(((((..((((((....))))))..)))))----------..((((((....((((((.(((((..(..(......)-..)..))-)))-((......)).....)))))))))))).. ( -26.70, z-score = -2.12, R) >droYak2.chrX 9789000 109 - 21770863 AUCGUUCGUCGCCAUCAGUGGCGAAAAGCGA----------CGAAAAGCUAAUAUGUAUAUAAAAAUGUUUUUCUUUG-UCCCCCCCUUUAGCGCCAAAGCAGCGAGCACAUGCUUUUAU .(((((..((((((....))))))..)))))----------.(((((((..................)))))))....-............((((....)).))((((....)))).... ( -25.87, z-score = -1.84, R) >droEre2.scaffold_4690 9593628 118 + 18748788 AUCGUUCGUCGCCAUCAGUGGCGAAAAGCGAGGAAAAGCGACGAAAAGCUAAUAUGUAUAUAAAAAUGUUUUCCUUUG-UCCCCCU-UCAAGCGCCAAAGCACCGGGCACAUGCUUUUAU .(((((..((((((....))))))..)))))..((((((((((((.....(((((..........)))))....))))-)).(((.-....((......))...))).....)))))).. ( -31.10, z-score = -1.55, R) >consensus AUCGUUCGUCGCCAUCAGUGGCGAAAAGCGA__________CGAAAAGCUAAUAUGUGUAUAAAAAUGUUUUCCUUUG_UCCCCCU_UUAAGCGCCAAAGCACCGAGCACAUGCUUUUAU .(((((..((((((....))))))..)))))............((((((.(((((..........))))).....................((......))...........)))))).. (-21.62 = -21.62 + -0.00)

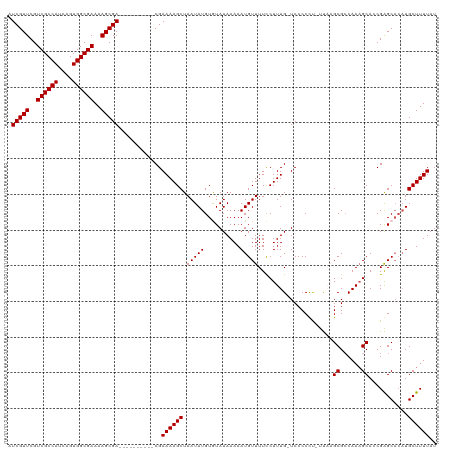
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:01:01 2011