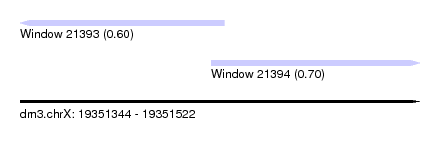
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,351,344 – 19,351,522 |
| Length | 178 |
| Max. P | 0.695781 |

| Location | 19,351,344 – 19,351,435 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 86.84 |
| Shannon entropy | 0.17888 |
| G+C content | 0.56532 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -27.34 |
| Energy contribution | -26.90 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.600043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19351344 91 - 22422827 GAAGCGGGAAGUUGCAAAGUUGAAGAGUGAGCCGGUGGGCAAGUGGGGAAUCCCCGUCCAUCAGCGCUUCAGCAUGCAUUGUGGCAUAGUG ...((.(.((..((((..(((((((.....((.(((((((....((((...))))))))))).)).))))))).)))))).).))...... ( -32.90, z-score = -1.90, R) >droSim1.chrX 14968316 78 - 17042790 -------GAAGUUGCAAAGUUGGAGAGUGAGCCGGUGGGCAAGUGGGGAAUCCCC-UCCAUCAGCGCUUCAGCAUGCAUAGUGCCA----- -------.....((((..(((((((.....((.((((((.....((((...))))-)))))).)).))))))).))))........----- ( -25.50, z-score = -0.42, R) >droSec1.super_8 1634953 84 - 3762037 -AAGUGGGAAGUUGCAAAGUUGGAGAGUGAGCCGGUGGGCAAGUGGGGAAUCCCC-UCCAUCAGCGCUCCAGCAUGCAUAGUGGCA----- -..((.(.....((((..(((((((.....((.((((((.....((((...))))-)))))).)).))))))).))))...).)).----- ( -28.90, z-score = -0.55, R) >consensus _AAG_GGGAAGUUGCAAAGUUGGAGAGUGAGCCGGUGGGCAAGUGGGGAAUCCCC_UCCAUCAGCGCUUCAGCAUGCAUAGUGGCA_____ ............((((..(((((((.....((.((((((.....((((...)))).)))))).)).))))))).))))............. (-27.34 = -26.90 + -0.44)


| Location | 19,351,429 – 19,351,522 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.91 |
| Shannon entropy | 0.40706 |
| G+C content | 0.60387 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -17.53 |
| Energy contribution | -18.78 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19351429 93 + 22422827 CGCUUCCCACUUCCCACUUGGCUGCGCCCGAAGUGGAGCAAAUUGAAAAGGG---GC--GUGGCCAC--GUGGCAGGUGGUGGACGUAAGGCAGAGGCAG .(((((((((((.((((.((((..(((((....(.((.....)).)....))---))--)..)))).--)))).))))))....(....)...))))).. ( -44.80, z-score = -2.96, R) >droAna3.scaffold_13417 4352067 89 - 6960332 -------CGCCCGCCACU-GCCCACAUU--GAGUGCAGAAAAUUGAAAAGGGCGUGCCAGUGGCAGCUGGUGACUGG-GGUGAAAGUAAGGCAAAAGUCG -------(((((((((((-((((...((--.(((.......))).))..)))).....))))))....(....)..)-)))).......(((....))). ( -27.90, z-score = -0.18, R) >droSim1.chrX 14968394 87 + 17042790 ------CCACUUCCCACUUGGCUGCGCCCGAUGUGGAGCAAAUUGAAAAGGG---GC--GUGGCCAC--GUGGCAGGUGGCGGACGCAAGGCGAAAGUCG ------((((((.((((.((((..(((((....(.((.....)).)....))---))--)..)))).--)))).)))))).........(((....))). ( -42.50, z-score = -3.08, R) >droSec1.super_8 1635032 92 + 3762037 -CACUUCCACUUCCCACUUGGCUGCGACCGAUGUGGAGCAAAUUGAAAAGGG---GC--GUGGCCAC--GUGGCAGGUGGCGGACGUAAGGCGAAAGUCG -..((.((((((.((((.((((..((.((....(.((.....)).)...)).---.)--)..)))).--)))).)))))).))......(((....))). ( -38.30, z-score = -2.23, R) >consensus ______CCACUUCCCACUUGGCUGCGCCCGAAGUGGAGCAAAUUGAAAAGGG___GC__GUGGCCAC__GUGGCAGGUGGCGGACGUAAGGCAAAAGUCG ......((((((.((((.(((((((..................................)))))))...)))).)))))).........(((....))). (-17.53 = -18.78 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:00:55 2011